Dynamic protein complex recognition method
A technology for protein complexes and identification methods, applied in proteomics, biological systems, biostatistics, etc., which can solve the problem that static PPI data cannot provide biological results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
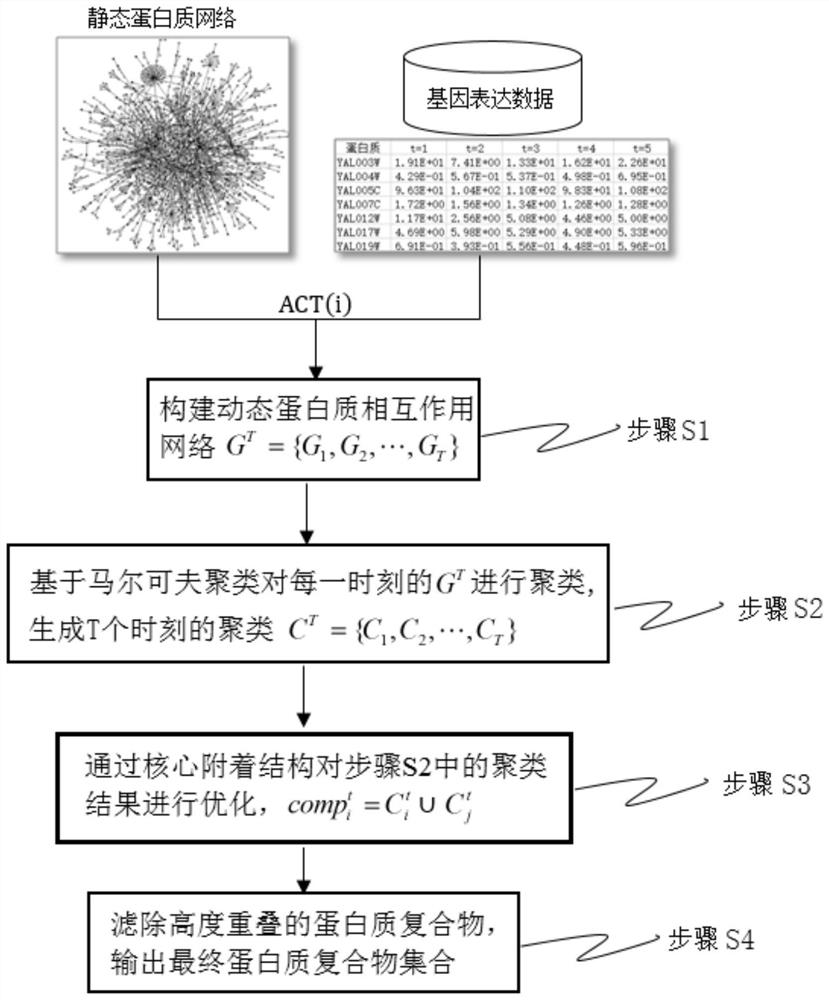
[0058] Further as a preferred embodiment of the present invention, the step S1 is specifically:
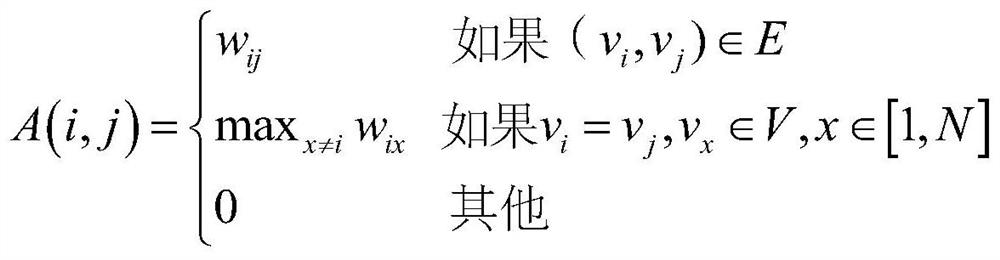
[0059] S11, the static PPI network is modeled as an undirected graph G=(V, E), wherein V represents a collection of protein nodes, and E represents a collection of edges between protein nodes;
[0060] S12. Determine the activity of the protein based on the gene expression data, and express the gene expression data of N proteins at T time points as an N×T matrix H; at time point t, assume that the gene expression value of a protein i is greater than or is equal to its activation threshold ACT(i), then the protein i is considered active; the expression of its activation threshold ACT(i) is as follows:
[0061] ACT(i)=u(i)+3σ(i)(1-F(i))
[0062] in, represents the average value of protein i from time 1 to T, σ(i) represents the standard deviation of protein i from time 1 to T; F(i)=1 / (1+σ 2 (i)) is a weight equation, which reflects the fluctuation of protein i expression value; ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com