Prawn growth related SNP markers and application thereof in breeding
A shrimp and marker technology, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, biochemical equipment and methods, etc., can solve the problems of small number of SNP markers, unclear analysis of growth traits, and inability to finely position genes, etc., to improve Breeding progress, accelerated genetic selection progress, and the effect of high breeding accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
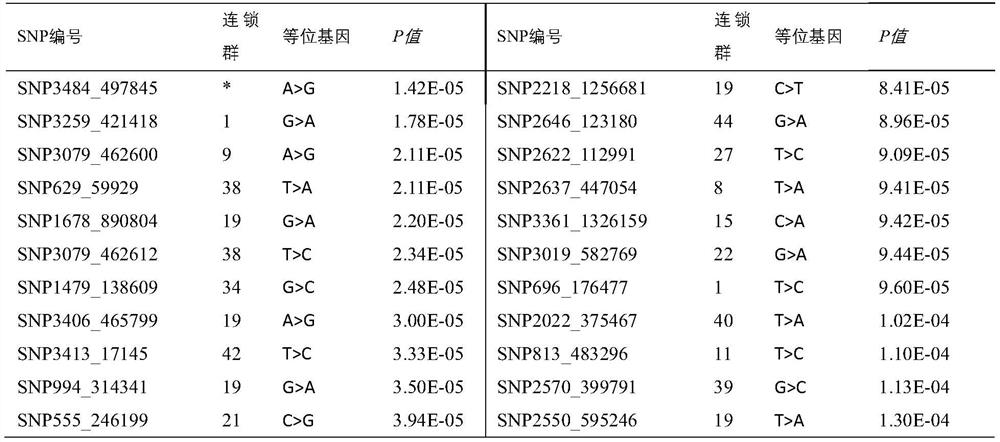
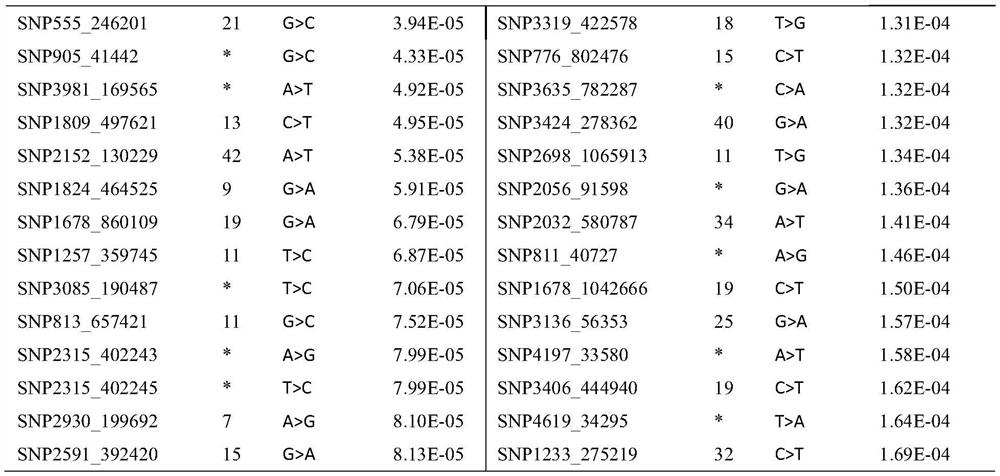
[0031] Embodiment 1: Obtaining of a group of SNP markers related to the growth of prawns
[0032] (1) Construction and trait determination of shrimp analysis population
[0033] The shrimp population used for analysis (Litopenaeus vannamei) was established in Hainan Guangtai Ocean Breeding Co., Ltd. in 2015. This population contains 13 full-sib families of shrimp. The establishment periods of these families are basically similar. Different families were cultured separately. After the family grew to about 3cm, 50 individuals were randomly selected from each family for VIE fluorescent labeling (NMT Company, USA), and the labeled shrimp were cultured under the same conditions. After 3 months of breeding, random selection The body weight of 200 individuals was measured, and the sex of the individuals was recorded.
[0034] (2) Sample DNA extraction
[0035] Use the Plant Genome Extraction Kit (Tiangen, Beijing) to extract the muscle tissue DNA of the above 200 individuals. For t...
Embodiment 2
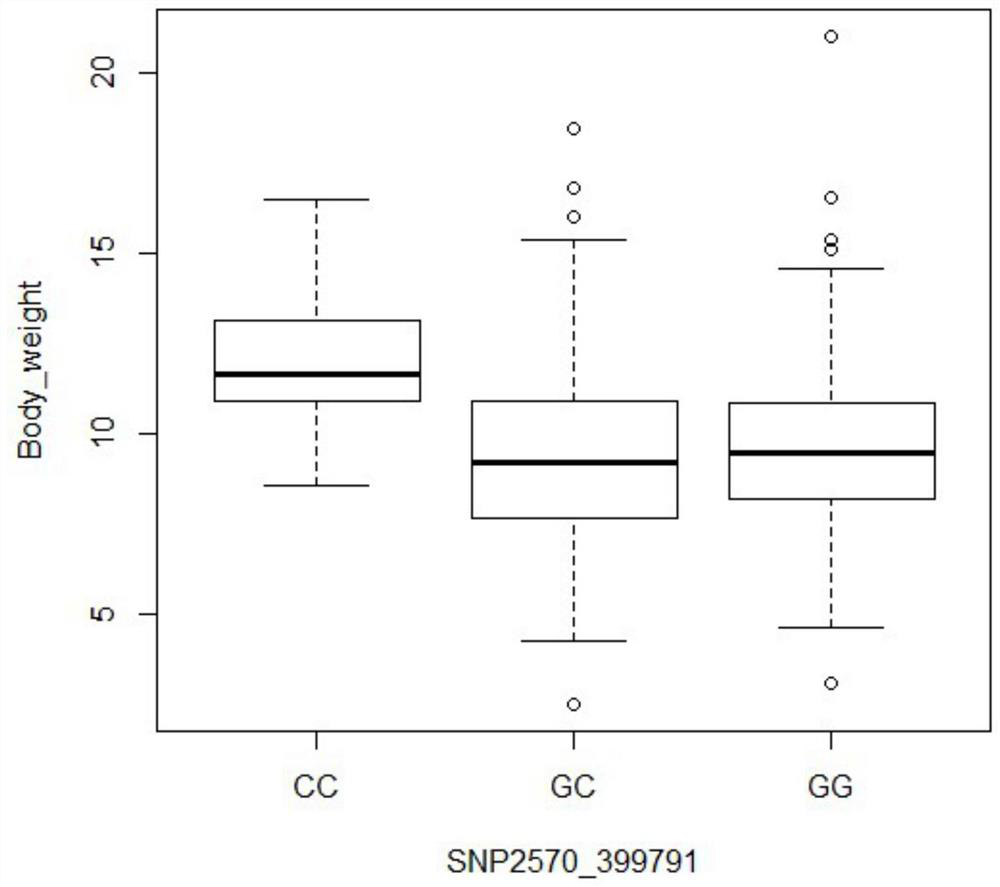
[0085] Example 2: Application of SNP2570_399791 marker in breeding
[0086] (1) Sources of breeding materials
[0087] The group used for analysis (Litopenaeus vannamei) was constructed at Hainan Guangshun Taipu Marine Breeding Co., Ltd. This group is a mixture of multiple families with the same breeding environment. After 3 months of breeding, 300 individuals were randomly selected , take the last pair of swimming feet for DNA extraction.
[0088] (2) Sample DNA extraction
[0089] The plant genome extraction kit (Tiangen, Beijing) was used to extract DNA from the above 300 individual tissues. For specific operation steps, refer to the kit instruction manual. The concentration of each individual DNA was measured using a nucleic acid concentration analyzer Nanodrop 1000, and detected by agarose gel electrophoresis. DNA integrity. (3) Marker typing of SNP2570_399791
[0090] Use primers to amplify the target sequence amplified by SNP2570F: CGAAATGCGAATCAGCCTTGA and SNP2570R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com