Recombinant parainfluenza virus type 5 for expressing double report tags
A parainfluenza and virus technology, applied in the field of recombinant parainfluenza virus type 5
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment 1, construction of rPIV5-GN cDNA clone
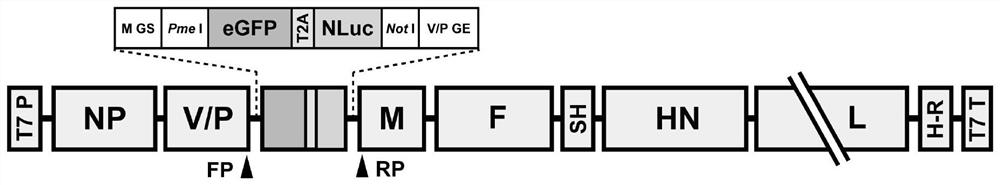
[0033] In this embodiment, the eGFP-T2A-NLuc fusion protein sequence is first constructed, the upstream includes the M gene start sequence (M GS) and the Pme I restriction site, and the downstream includes the Not I restriction site and the V / P gene termination sequence (V / P GE). eGFP-T2A-NLuc fusion protein sequence as figure 1 , the arrow indicates the self-cleavage site. On this basis, the recombinant PIV5 nucleic acid fragment rPIV5-GN cDNA clone used to rescue co-expression of eGFP and NLuc was constructed. The fragment is distributed in "5'-NP-V / P-eGFP-T2A-NLuc-M-F-SH-HN-L-3'" ( figure 2 ). Among them, NP, V / P, M, F, SH, HN, and L genes are related genes of PIV5 strain (GenBank: NC_006430.1); eGFP is enhanced green fluorescent protein ORF; T2A is a self-cleaving peptide sequence; NLuc ORF for NanoLuc luciferase.
[0034] The nucleotide sequence of the rPIV5-GN cDNA clone constructed in this example is SEQ...
Embodiment 2
[0036] Example 2, Rescue of rPIV5-GN
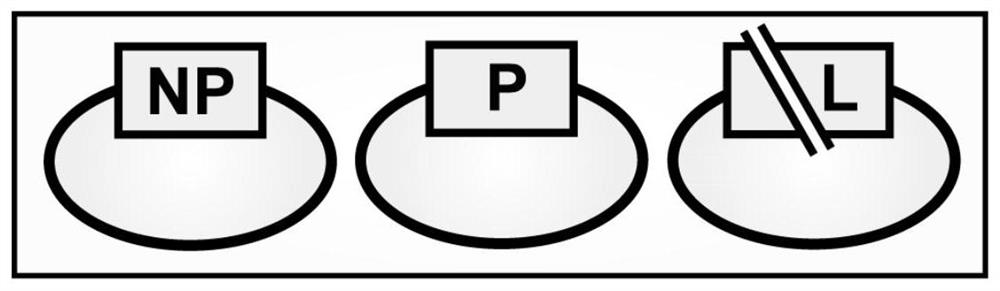
[0037] Resuscitate BSR-T7 / 5 cells, passage twice in DMEM medium, DMEM medium contains penicillin (100U / mL), streptomycin (100μg / mL), amphotericin B (0.25μg / mL), G418 (500μg / mL) And 10% FBS (fetal bovine serum). The day before transfection, BSR-T7 / 5 cells were inoculated in 12-well plates, and the next day when the cells reached 70% confluence, the rPIV5-GN cDNA clone plasmid and three auxiliary plasmids ( image 3) co-transfected BSR-T7 / 5 cells, and the transfection amounts of the four were 2 μg, 1 μg, 0.5 μg, and 0.5 μg per well, respectively. Solution I was prepared by adding 75 μL Opti-MEM and 8 μL Lipofectamine2000 to each well. Then add 75 μL Opti-MEM and 4 μg plasmid to each well to prepare solution II. Gently mix solutions I and II, and place at room temperature for 15 minutes to form liposome-DNA complexes. Add the liposome-DNA mixture dropwise to the cell wells. Place the 12-well plate in CO 2 After transfection in the incu...
Embodiment 3
[0038] Embodiment 3, identification of rPIV5-GN
[0039] The rescued rPIV5-GN was continuously blindly passed to the seventh passage in MDBK cells. With the blind passage of the virus, eGFP reached the desired expression level, but no cytopathic changes were observed throughout the passaging process ( Image 6 ). At the same time, 48 hours after the seventh-generation rPIV5-GN infected cells, NLuc could also be highly expressed ( Figure 7 ). The 7th passage virus solution was frozen and thawed once, and the supernatant was collected to extract total RNA, which was used for RT-PCR detection of the virus. Upstream primer sequence: 5'-GCAATCATCCGCAGTGCAATTTGA-3', downstream primer sequence: 5'-TGCGGGAATGCTGATGGATGGCAT-3', preamplified 1578bp fragment. The RT-PCR reaction adopts HighFidelity One Step RT-PCR Kit (Takara), and the reaction conditions are set as follows: 45°C for 10min, 94°C for 2min, and 30 cycles: 98°C (10s), 55°C (15s), 68°C ( 16s). In order to eliminate re...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap