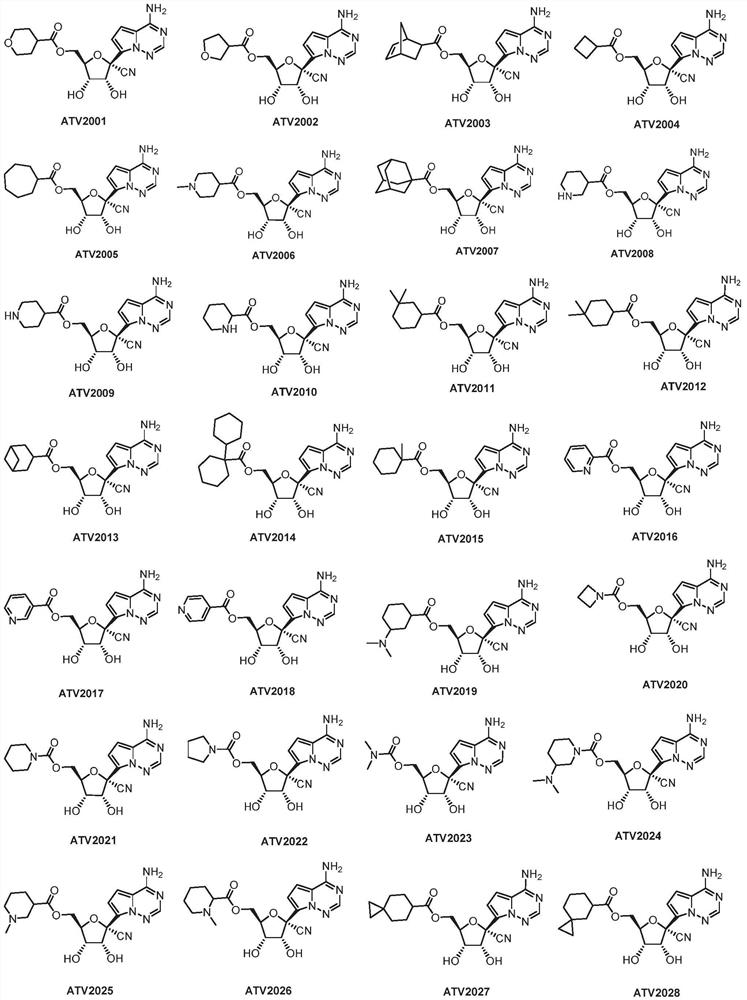
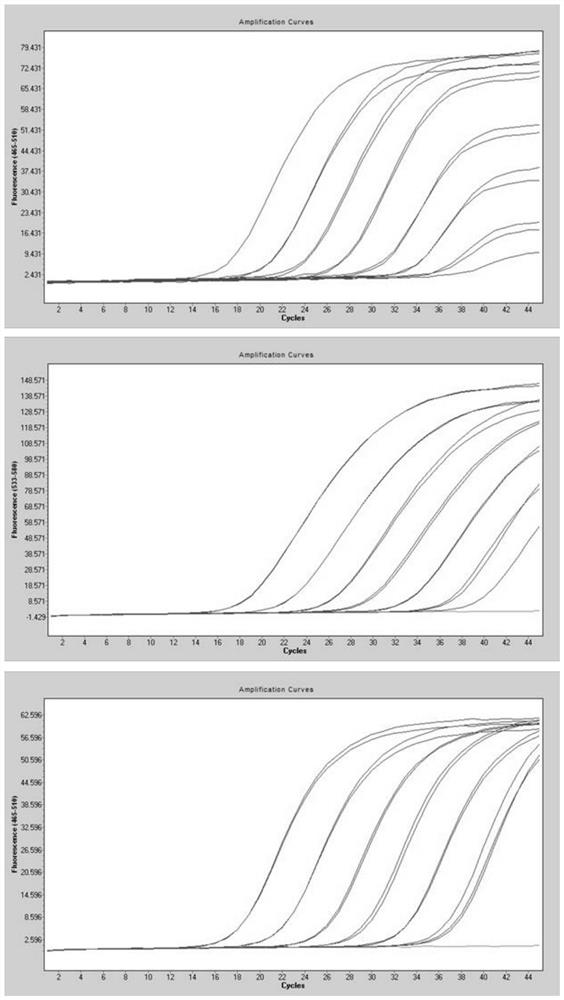
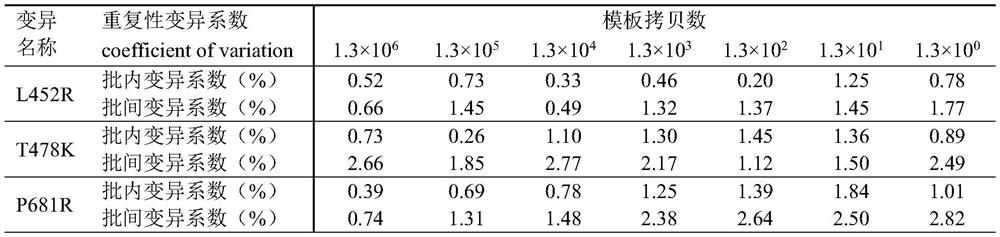
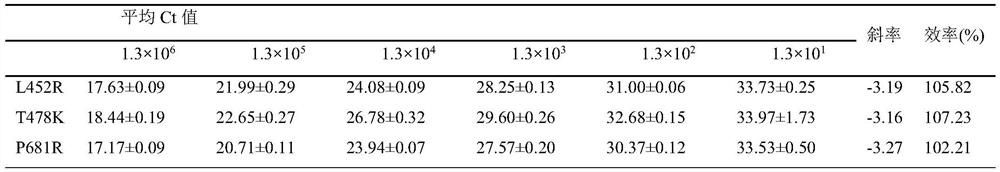
Patents
Literature
34 results about "Variant virus" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A variant virus is an influenza virus that normally circulates and causes illness in pigs, but once a human is infected, it is called a "variant" influenza virus.
Porcine pseudorabies virus (PRV) variant PRV-ZJ01 and application thereof
ActiveCN103627678AImprove securityImproving immunogenicityMicroorganism based processesAntiviralsRabiesEngineering
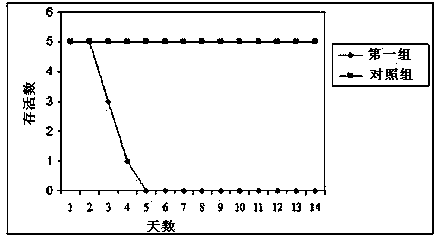
The invention relates to the technical field of porcine pseudorabies viruses (PRVs) and in particular relates to a porcine PRV variant PRV-ZJ01 with collection number of CGMCCNo.8170 and an application of the porcine PRV variant PRV-ZJ01 in preparation of vaccines. The porcine PRV variant PRV-ZJ01 has the beneficial effects that a water-soluble inactivated vaccine is prepared by adopting a PRV-ZJ01 variant virus solution and is subjected to a swine immune protection test with live vaccines of Bartha-K61, Bucharest and HB-98 strains and the results show that the inactivated vaccine of the ZJ01 strain has relatively high safety and has the immune protection efficiency obviously higher than that of immunity groups of the live vaccines of the Bartha-K61, Bucharest and HB-98 strains, and the live vaccines of the Bartha-K61, Bucharest and HB-98 strains can not provide full protection for the ZJ01 very virulent strain; the inactivated vaccine of ZJ01 has relatively good immune protection effects on the PRV variant and the traditional strains; infected with 10<6.0>TCID50 (Tissue culture infectious dose 50) / ml nasal drops of the PRV-ZJ01 variant, all the 85-day-old non-immune swine can become ill and die; results prove that the virulence of the virus strain is obviously enhanced, the antigenicity is varied and the virus strain has relatively good immunogenicity after being inactivated and can be used for research and development of the vaccine of the virus strain and the diagnostic methods.
Owner:NANJING AGRICULTURAL UNIVERSITY
gE- and gI-deleted porcine pseudorabies virus variant strain and use thereof
The invention relates to the technical field of porcine pseudorabies viruses and especially relates to a gE- and gI-deleted porcine pseudorabies virus variant strain PRV-ZJ011G and a use thereof. The gE- and gI-deleted porcine pseudorabies virus variant strain PRV-ZJ011G has the accession number of CGMCC No.7957. The invention discloses the use of the gE- and gI-deleted porcine pseudorabies virus variant strain PRV-ZJ011G in vaccine preparation. After the New Zealand big white rabbit is inoculated with the 106.0TCID50 recombinant viruses, clinical symptoms such as pruritus are not caused. An oil-in-water inactivated vaccine prepared from the gE- and gI-deleted porcine pseudorabies virus variant strain PRV-ZJ011G is injected into a piglet and after four weeks, the BELISA antibody is produced but the gE antibody does not exist, and the immunization protection efficiency is 100%. After immunization on sows, the piglets produced by the sows get immunization protection and the efficiency of PRV variant virus and traditional virus immunization protection is 100%. It is proved that the ZJ011G recombinant virus has good immunogenicity and can be used for vaccine preparation.
Owner:JIANGSU NANNONG HI TECH
Application of compound to treatment of SARS-CoV-2 infection
InactiveCN111991401AInhibition of replicationHigh activityAntiviralsRespiratory disorderCytopathic effectSide effect
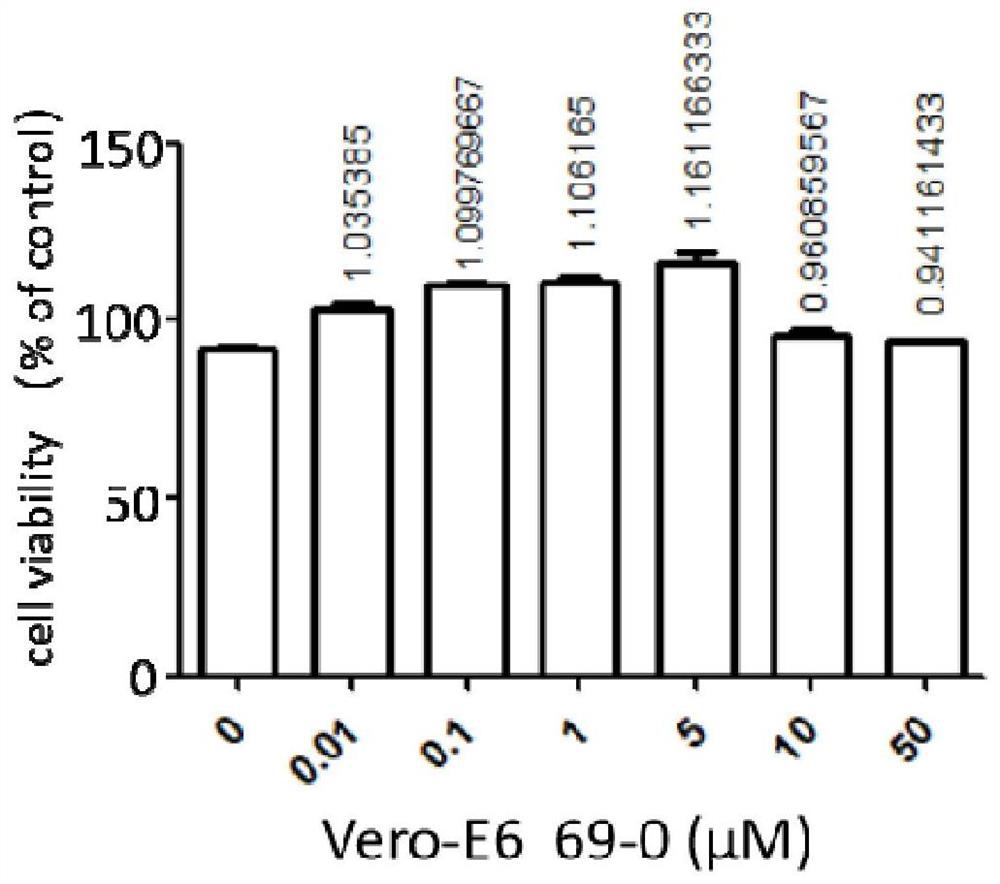
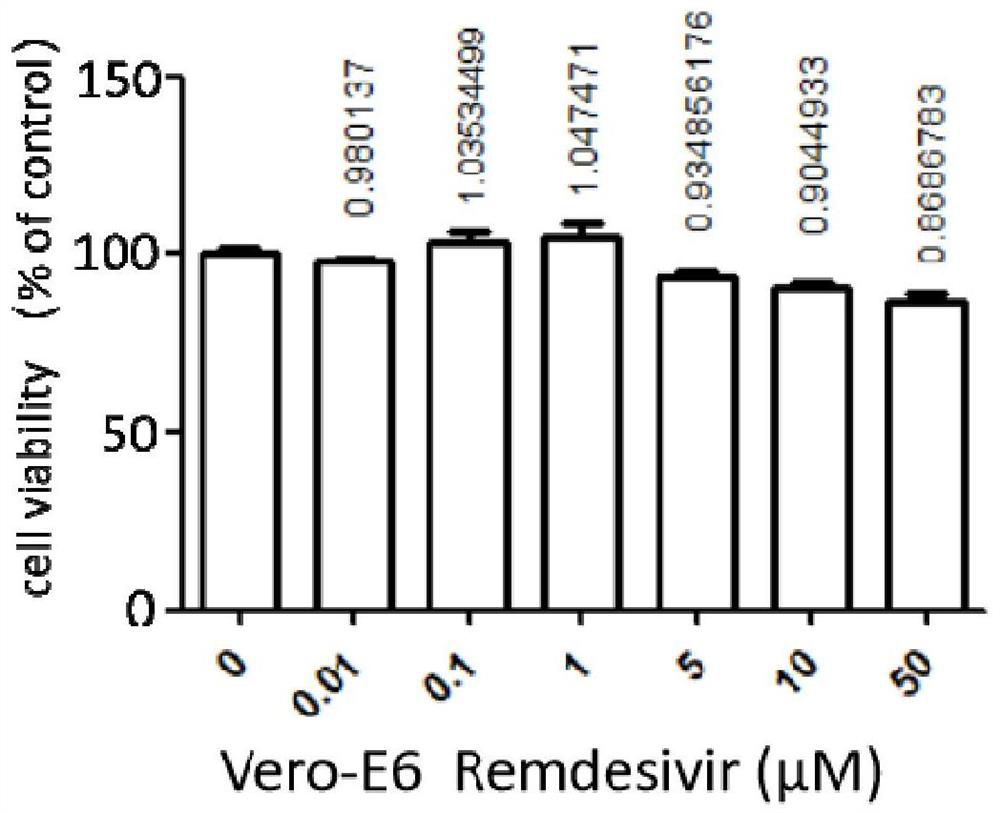
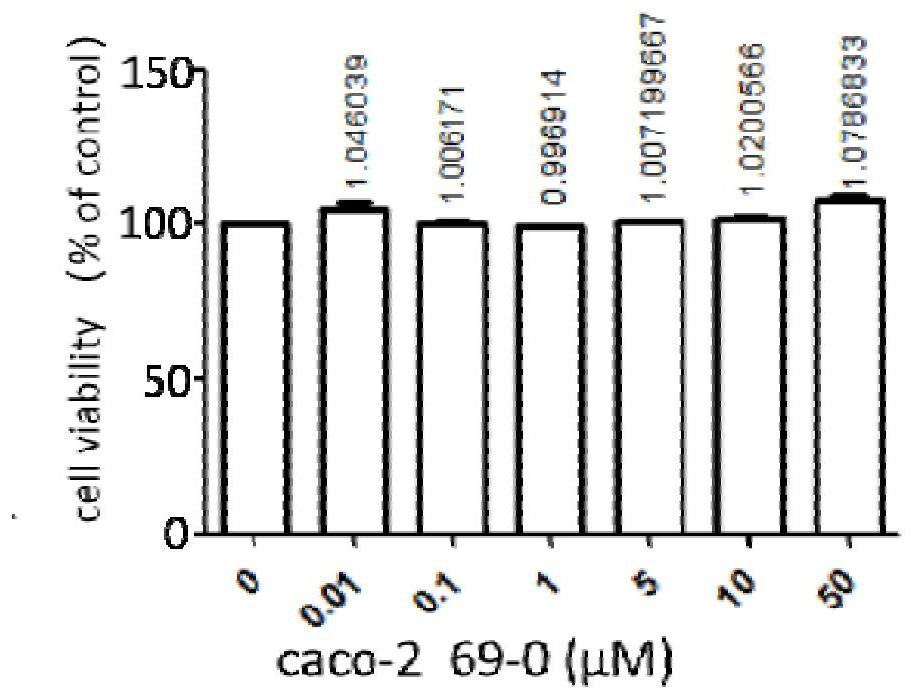
The invention provides an application of a compound 69-0 and / or tautomers and / or pharmaceutically acceptable salts thereof to prevention, alleviation and / or treatment of COVID19 / SARS-CoV-2. Products containing the compounds can effectively inhibit replication or reproduction of SARS-CoV-2 or homologous variant viruses of SARS-CoV-2 and the cytopathic effect produced by SARS-CoV-2. The compound 69-0 has good curative effect, good safety and low toxic and side effects. The invention also provides combined therapeutic pharmaceutical composition. The combined therapeutic pharmaceutical compositioncan effectively inhibit proliferation of SARS-CoV-2 in cells, and has good curative effect, low safety and low toxic and side effects, can reduce drug resistance of viruses and has obvious synergistic effect.
Owner:SOUTH UNIVERSITY OF SCIENCE AND TECHNOLOGY OF CHINA +1
Methods of treating viral infection
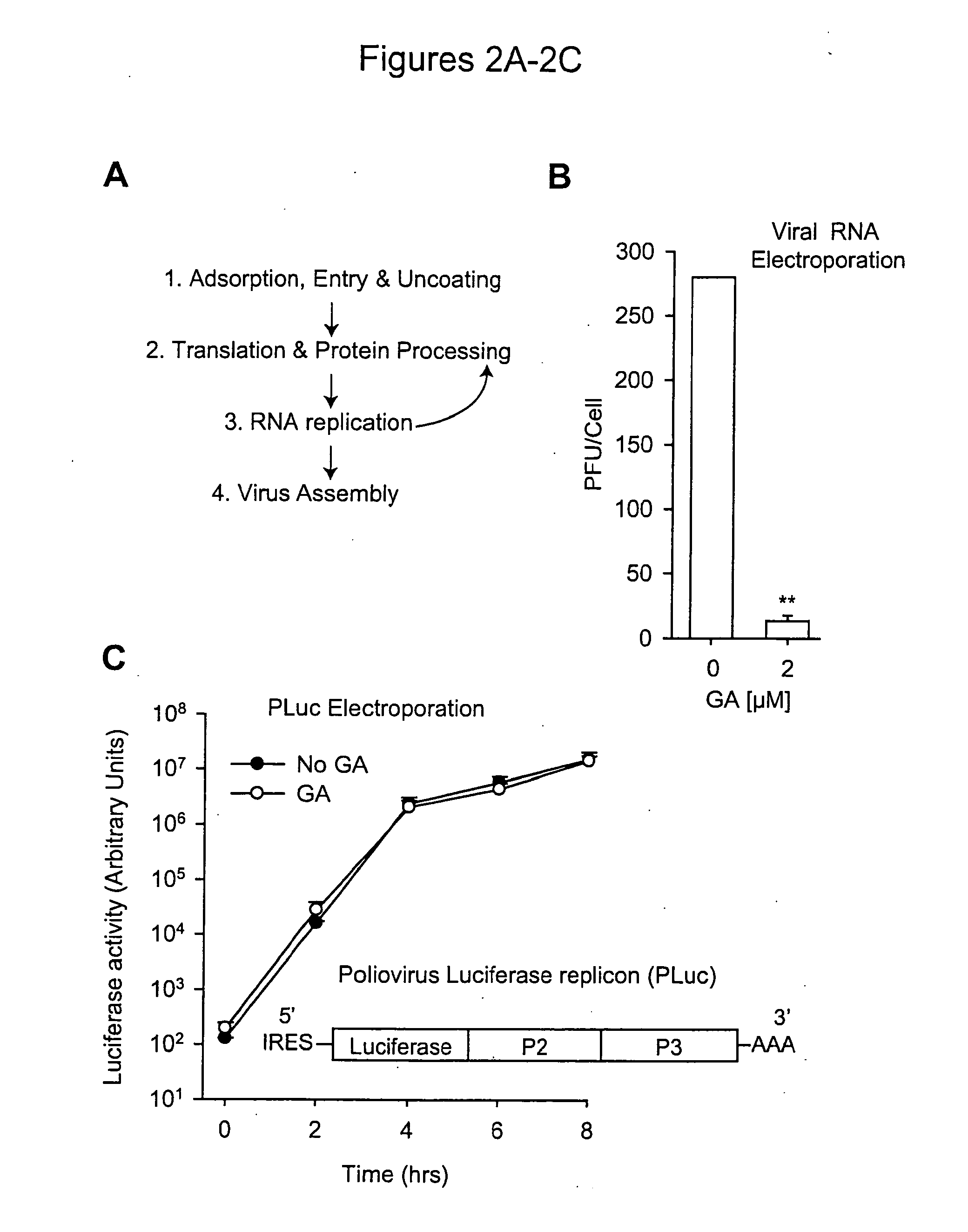
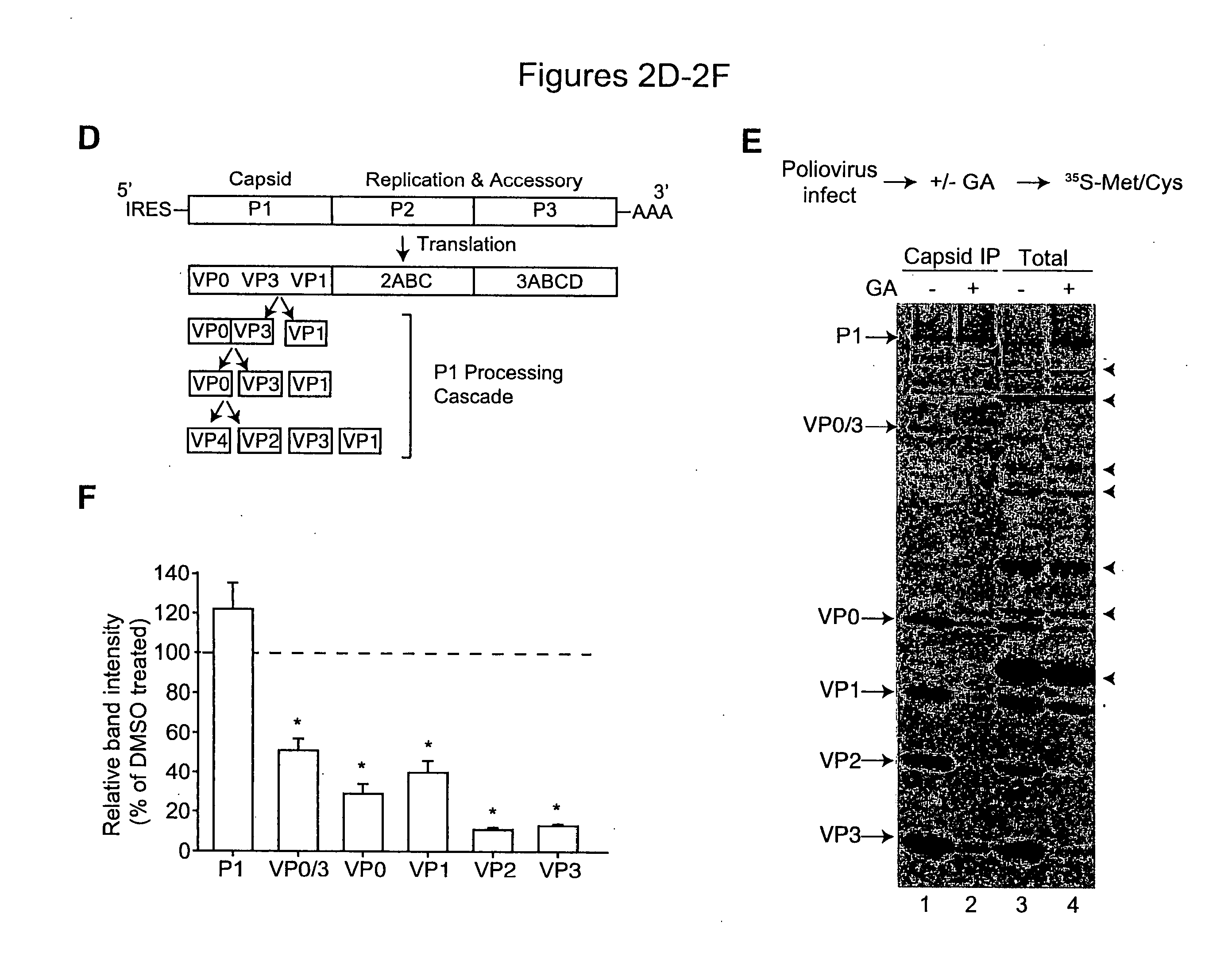
The present invention provides methods of treating an RNA viral infection, generally involving administering an agent that reduces the activity of a host cell protein required for maturation of a viral protein, where the emergence of variant virus resistant to the agent is reduced. The present invention further provides combination therapies for viral infection, involving administration of two or more agents that reduce the activity of a host cell protein required for maturation of a viral protein.
Owner:RGT UNIV OF CALIFORNIA +1
Immunogenic composition comprising an influenza virus with a temperature sensitive PB2 mutation
InactiveUS6843996B1Appropriate level of attenuationGenetic stabilitySsRNA viruses negative-senseVirus peptidesVariant virusProphylactic treatment
Recombinant PB2 variant influenza viruses, RNA, cDNA and vectors are provided. Also provided are immunogenic compositions containing the variant viruses, methods of producing such viruses and methods for the prophylactic treatment of influenza in humans.
Owner:MEDIMMUNE LLC
Single clone antibody of antimutagen hepatitis B virus surface antigen
InactiveCN1680581AAvoid losing toQuality assuranceImmunoglobulins against virusesFused cellsAntigenVariant virus
A process of preparing single-colon antibody resisting surface antigen of hepatitis B virus mutant from CGMCCHBSP2. The single-colon antibody can detect 14 types hepatitis B virus containing mutant and wild strain and replace HBsAb in fluorogence diagnostic kit. It can avoid mistake induced by normal agent and be used widely.
Owner:徐钧
Nucleoside compound and application thereof
PendingCN114292272AInhibition of replicationAntibacterial agentsOrganic chemistryCytopathic effectVariant virus
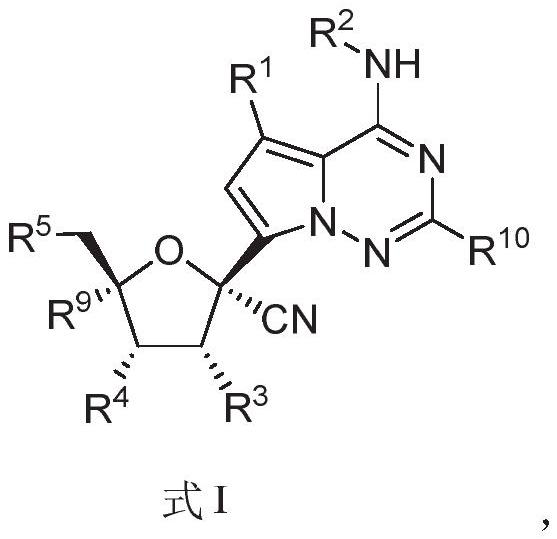
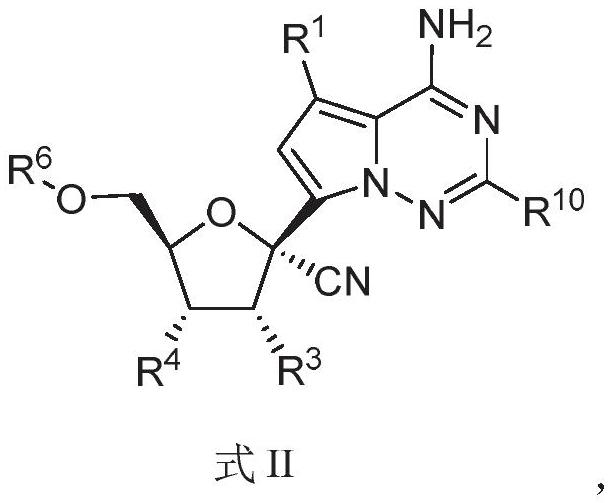
The invention relates to a nucleoside compound and application thereof, in particular to a compound shown in a formula I, a prodrug thereof and / or pharmaceutically acceptable salt thereof, and a preparation method, a composition and application thereof. The compound and the composition have the application of preventing, relieving and / or treating coronavirus infection, or replication or reproduction of homologous variant viruses of the coronavirus infection, and cytopathic effects generated by the coronavirus infection or the replication or reproduction of the homologous variant viruses of the coronavirus infection.
Owner:SOUTH UNIVERSITY OF SCIENCE AND TECHNOLOGY OF CHINA +1
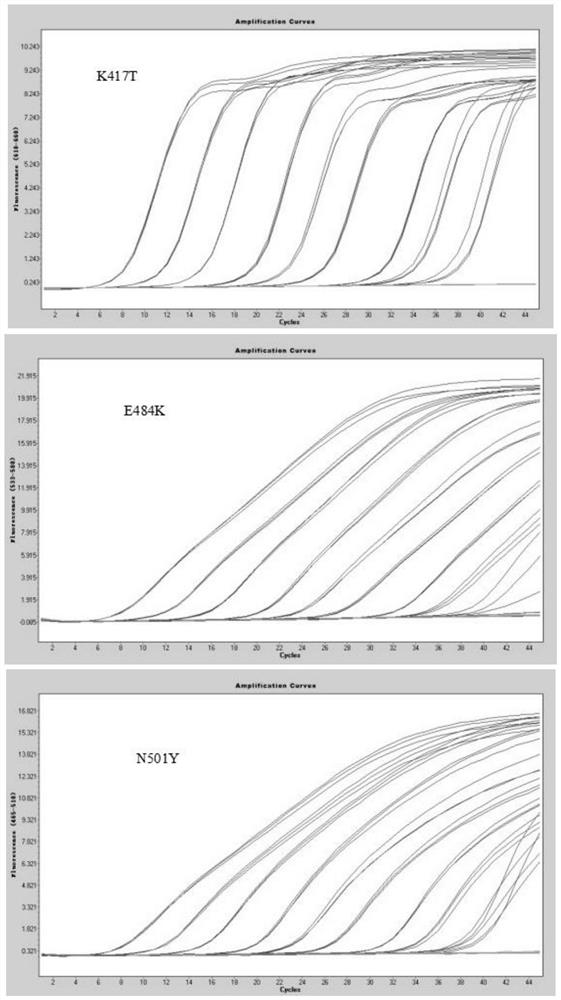
QRT-PCR (quantitative reverse transcription-polymerase chain reaction) method for identifying Indian variant of novel coronavirus
PendingCN113249525AReduce dependenceSave time for transshipmentMicrobiological testing/measurementAgainst vector-borne diseasesMutation detectionVariant virus
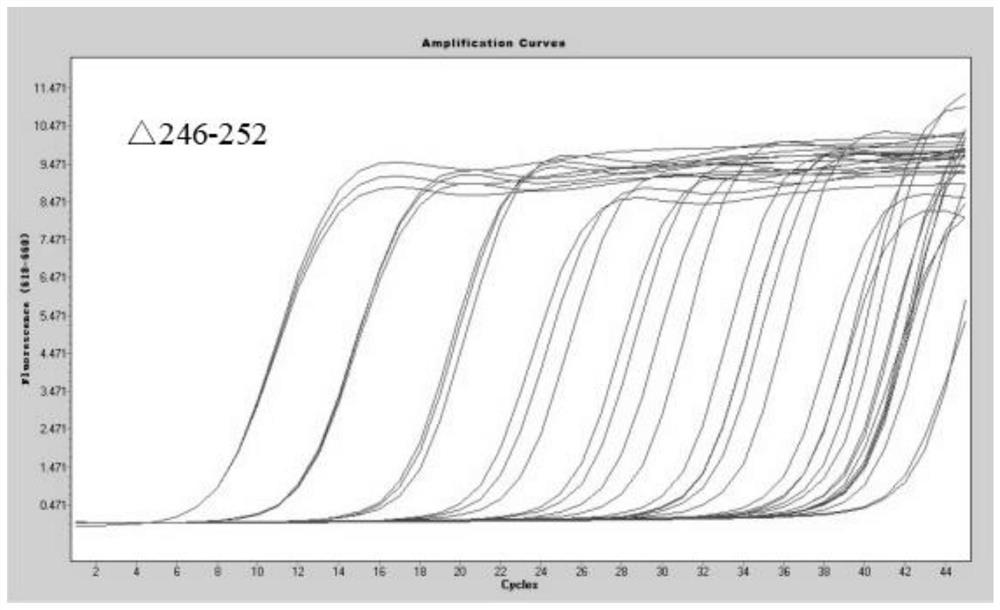
The invention belongs to the field of biotechnology, and particularly relates to a qRT-PCR method for identifying Indian variant of novel coronavirus. According to the method, in order to improve the detection sensitivity and specificity, a TaqMan probe is introduced; in order to reduce the cost, two pairs of primers are changed into one pair and a half of primers, that is, two upstream primers respectively target a mutation site and an original non-mutation site, and one downstream primer is shared by the two upstream primers. in order to improve the sensitivity of mutation detection, mutation is introduced to the third site in the 5' direction of the mutation site of the upstream mutation primer, and by reducing the matching degree of the mutation primer and non-mutated virus nucleic acid and the matching degree of the non-mutated primer and the mutation virus nucleic acid in the reaction system, the amplification curve of the mutation virus nucleic acid amplified by mutation primer in the reaction system is earlier than the amplification curve of the non-mutation nucleic acid, and in the same way, the amplification curve of the non-mutation nucleic acid amplified by the non-mutation primer is earlier than the amplification curve of the mutated nucleic acid.
Owner:SHANXI UNIV
Recombinant tryptophan mutants of influenza
InactiveUS6974686B2Lower Level RequirementsGenetic stabilitySsRNA viruses negative-senseSugar derivativesHuman influenzaTryptophan
Recombinant PB2 tryptophan variant influenza viruses, RNA, cDNA and vectors are provided. Also provided are immunogenic compositions containing the variant viruses, methods of producing such viruses and methods for the prophylactic treatment of influenza in humans.
Owner:MEDIMMUNE VACCINES
Detection method of new coronavirus SARS-CoV-2 antigen
ActiveCN113447467AStable dispersed stateStrong specificityBiological material analysisRaman scatteringSpecific immunityVariant virus
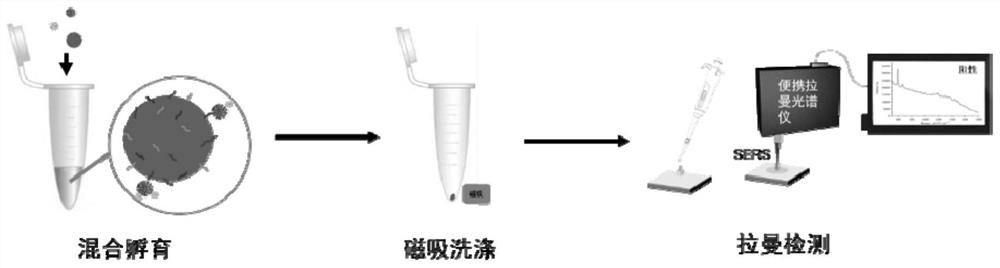
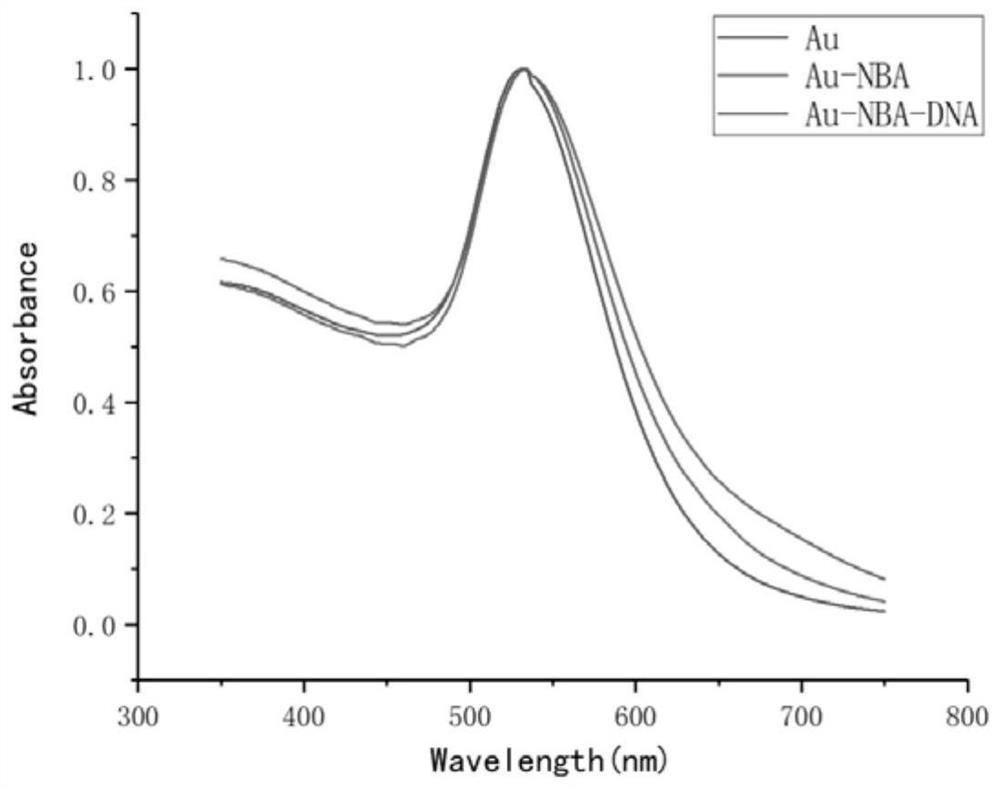
The invention discloses a detection method of a new coronavirus SARS-CoV-2 antigen, and belongs to the technical field of biology. The method comprises the following steps of capturing nanoparticles by virtue of new coronavirus SARS-CoV-2S protein immunomagnetic beads, carrying out specific immunization on the nanoparticles and S protein, and incubating with immune gold signal nanoparticles so as to construct a sandwich structure; detecting an SERS signal through a Raman spectrometer, specifically detecting the new coronavirus SARS-CoV-2 protein within 5 minutes, and specifically detecting the pseudovirus and variant pseudovirus of the SARS-CoV-2 and the pseudovirus diluted in human saliva. Through the above mode, nucleic acid extraction or antibody-dependent detection is not needed, meanwhile, the method has the advantages of being good in stability, higher in specificity and affinity, low in immunogenicity, easy to chemically modify, simple and convenient to operate and the like, the rapid detection of the new coronavirus SARS-CoV-2 antigen and the variant virus can be achieved within five minutes, and a condition is created for the early detection of the 2019SARS-CoV-2 pathogen.
Owner:杭州佑戈科技有限公司
Matrix protein mutated recombinant vesicular stomatitis virus serving as porcine vaccine vector
InactiveCN103768592AExperimental results directlyThe experimental results are accurateAntiviralsAntibody medical ingredientsArginineVariant virus
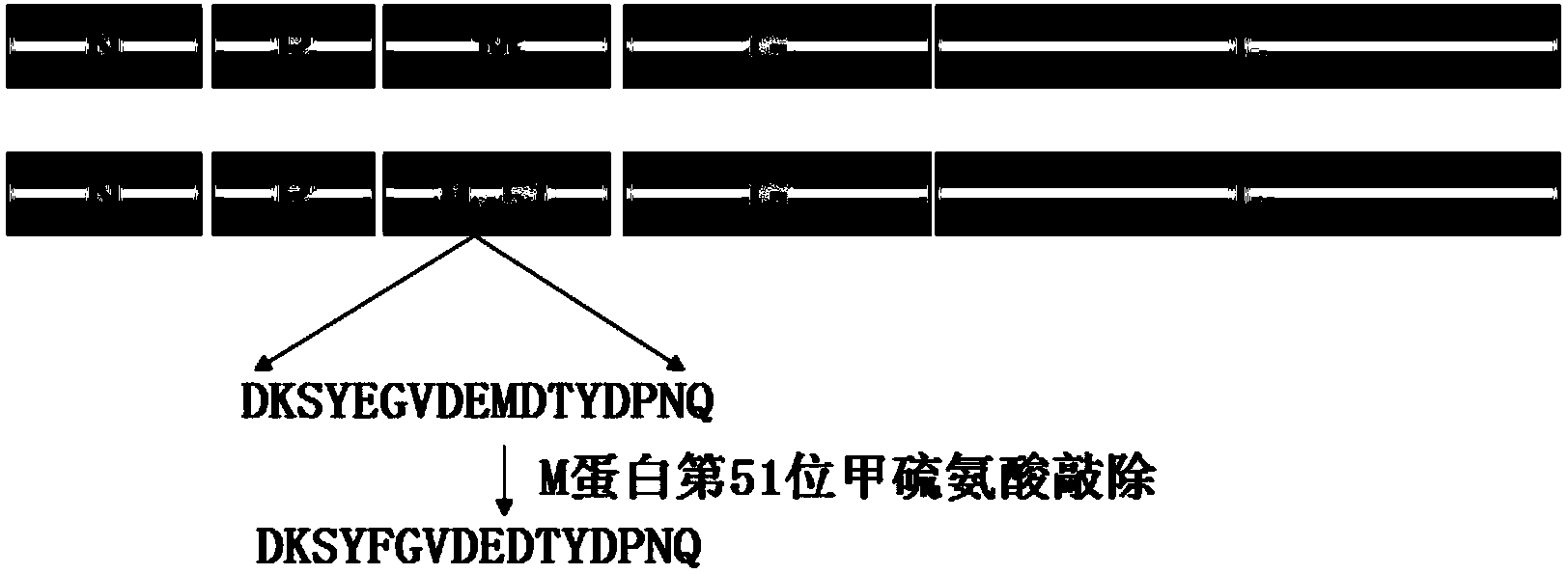
The invention discloses a matrix protein mutated recombinant vesicular stomatitis virus serving as a porcine vaccine vector. The vaccine vector is a recombinant virus VSV (vesicular stomatitis virus) delta M51. The invention also provides a kit for detecting the specific M antibody in porcine serum. The using method of the kit comprises the following steps: establishing an ELISA method by utilizing high-purity M protein prepared by recombinant expression to detect the level of the specific M antibody in porcine serum which is inoculated with VSV delta M51 virus. The invention firstly provides the variant virus strain (VSV delta M51) of which the 51st arginine of the matrix protein of an VSV critical virulence factor serving as a candidate VSV virus strain with application values, provides a systematic evaluation to the pathogenicity of VSV delta M51 on pigs for the first time at home and abroad, and provides basis to VSV delta M51 serving as the vaccine vector.
Owner:SHANGHAI JIAO TONG UNIV
Method for detecting COVID-19 based on mNGS and application of method
ActiveCN113337639AImprove reverse transcription efficiencyLow pollution rateMicrobiological testing/measurementLibrary creationNucleotideVariant virus
The invention provides a method for detecting COVID-19 based on mNGS and application of the method. According to the method, a multiple genome specific reverse transcription primer group of COVID-19 is designed and prepared, so that the reverse transcription efficiency of RNA of COVID-19 is improved, the aerosol pollution problem is reduced, and meanwhile, the enrichment capacity of different variant virus nucleotide sequences is improved.
Owner:天津金匙医学科技有限公司
Adenovirus vector recombinant new coronavirus B.1. 1.529 variant vaccine and application thereof
ActiveCN114395569AEfficient expressionSsRNA viruses positive-senseViral antigen ingredientsVector vaccineAntigen binding
The invention provides a novel coronavirus B.1. 1.529 variant vaccine taking a human type 5 replication-deficient adenovirus as a vector. The invention further provides a preparation method of the novel coronavirus B.1. 1.529 variant vaccine. On the premise that an expression protein main body is still novel coronavirus B.1. 1.529 variant spike protein, a recombinant virus vector vaccine prepared from a nucleotide sequence after experience optimization can effectively stimulate an organism to generate a binding antibody, a neutralizing antibody and cellular immune response aiming at the B.1. 1.529 variant virus after immunization, and has good immunogenicity.
Owner:ACADEMY OF MILITARY MEDICAL SCI
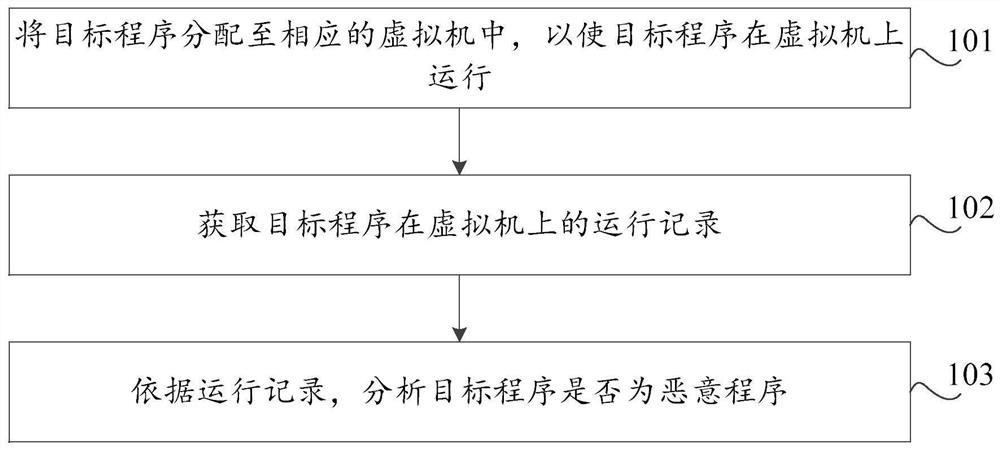
Malicious program detection method and device, storage medium and computer equipment
ActiveCN112580041AAffect securityAffect protection securityPlatform integrity maintainanceEnergy efficient computingSoftware engineeringVariant virus
The invention discloses a malicious program detection method and device, a storage medium and computer equipment, and the method comprises the steps: distributing a target program to a corresponding virtual machine, so as to enable the target program to run on the virtual machine; obtaining a running record of the target program on the virtual machine; and analyzing whether the target program is amalicious program or not according to the running record. By utilizing the virtual machine technology, the running record of the target program during execution in the virtual machine can be effectively obtained, the real computer environment is not influenced, the safety of the computer is protected, meanwhile, heuristic virus detection is carried out on the basis of the running record of the target program. The detection rate of unknown viruses such as novel viruses and variant viruses is favorably improved.
Owner:QI AN XIN SECURITY TECH ZHUHAI CO LTD +1
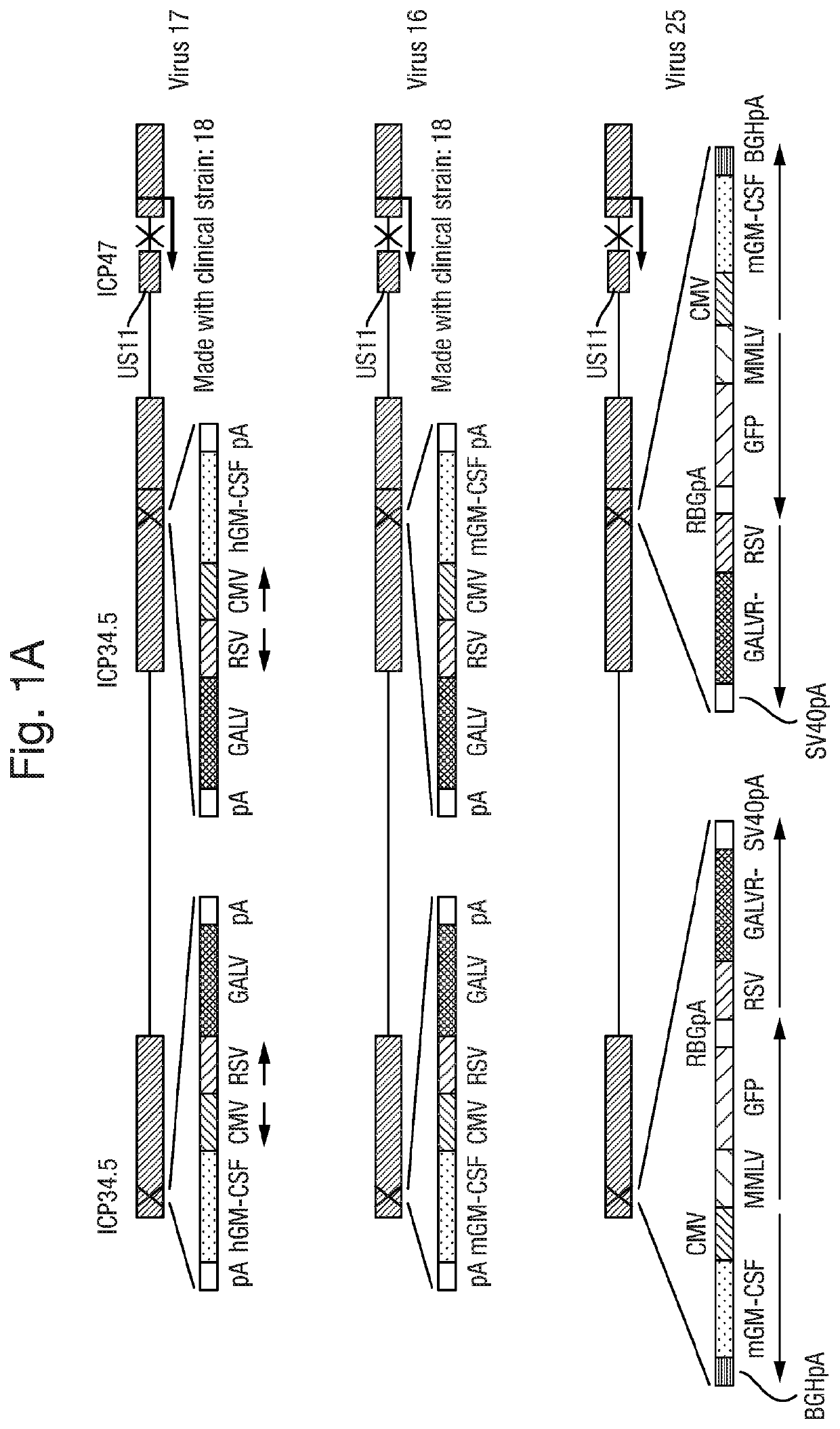
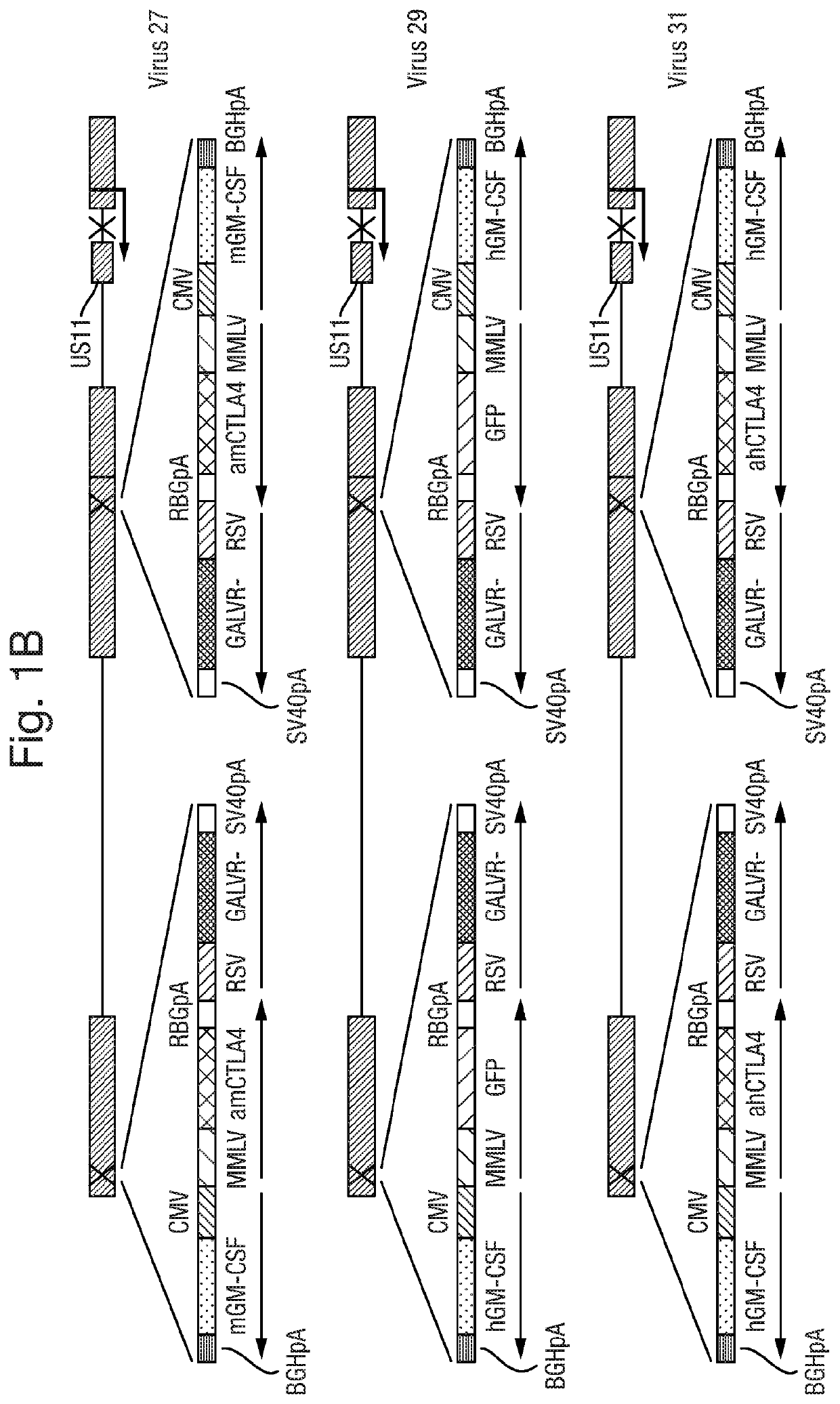
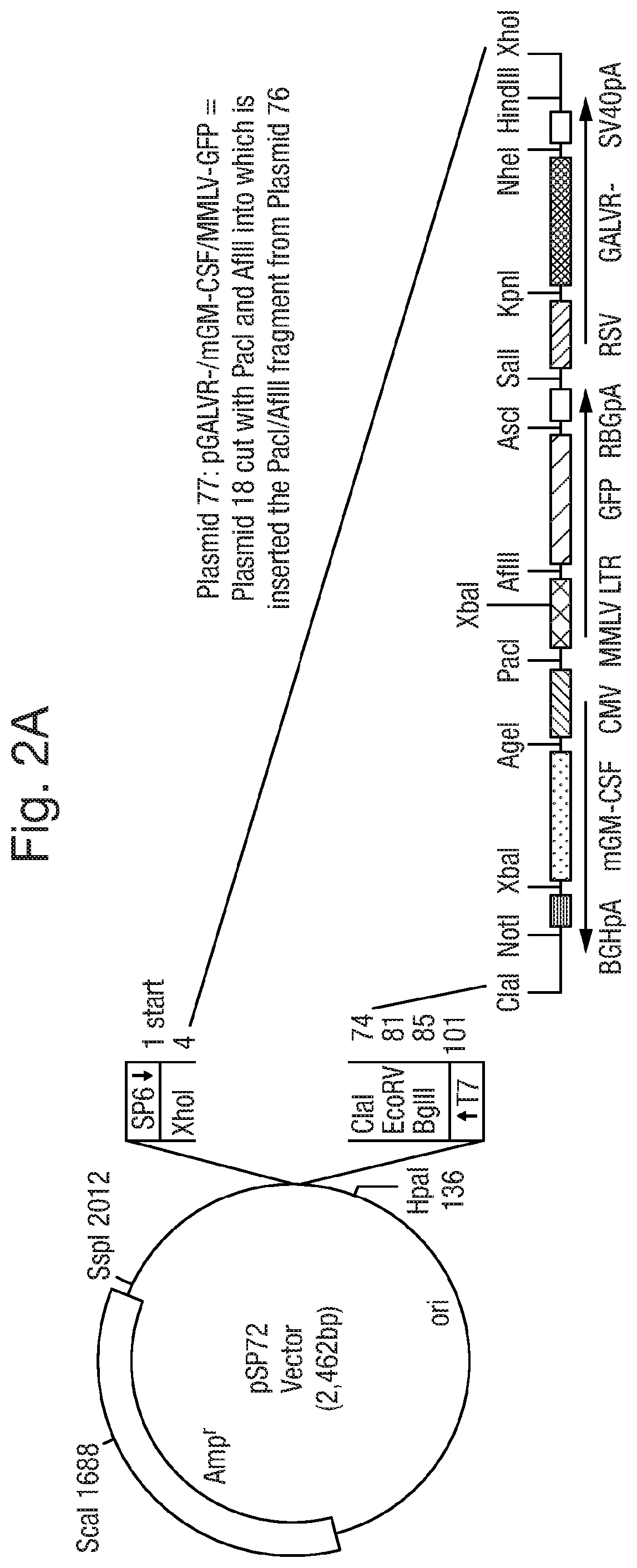
Altered virus
PendingUS20190343903A1Stimulate proliferation and maturationReduced amplificationImmunoglobulins against cell receptors/antigens/surface-determinantsAntibody ingredientsCTLA-4Antigen
The present invention relates to an oncolytic virus encoding a CTLA-4 inhibitor, such as an anti-CTLA-4 antibody, or an antigen binding fragment thereof.
Owner:REPLIMUNE
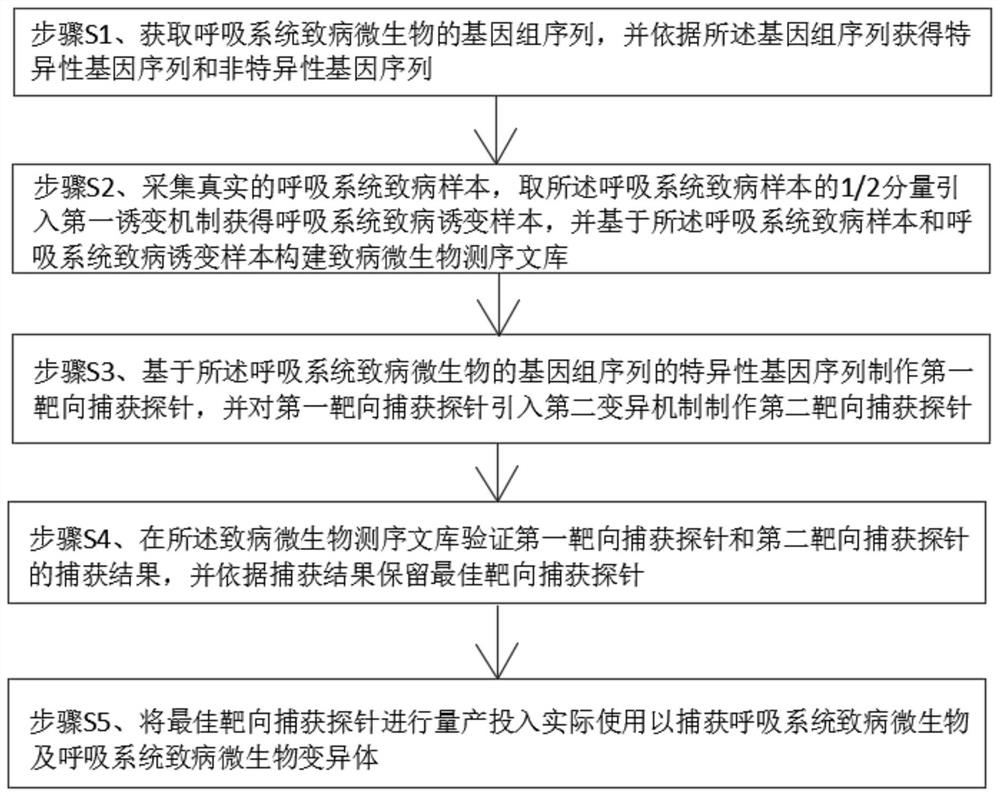
Targeted capture sequencing detection method for pathogenic microorganisms of respiratory system
InactiveCN113046478AIncrease gene sequence abundanceAvoid Sequencing ResultsMicrobiological testing/measurementDNA preparationPathogenic microorganismTarget capture
The invention discloses a targeted capture sequencing detection method for pathogenic microorganisms of a respiratory system. The method comprises the following steps of: S1, acquiring a genome sequence of the pathogenic microorganisms of the respiratory system, and acquiring a specific gene sequence and a non-specific gene sequence according to the genome sequence; and S2, collecting a real pathogenic sample of the respiratory system, introducing a first mutagenesis mechanism into a half component of the pathogenic sample of the respiratory system to obtain a pathogenic mutagenesis sample of the respiratory system, and constructing a pathogenic microorganism sequencing library based on the pathogenic sample of the respiratory system and the pathogenic mutagenesis sample of the respiratory system. According to the method, a mutation mechanism is introduced into the preparation of the pathogenic microorganism sequencing library to simulate a microorganism mutation process, so that the gene sequence abundance of the microorganisms is increased, a sequencing result of the mutated virus microorganisms is prevented from being false negative, and the sequencing accuracy is improved.
Owner:深圳人体密码基因科技有限公司
Porcine pseudorabies variant virus strain and application thereof
The invention provides a porcine pseudorabies variant virus strain and application thereof, and belongs to the technical field of virus molecular biology and genetic engineering. A virus separated from a PRV (Porcine Reproductive Virus) infected body in a pig farm is confirmed to belong to a PRV virulent virus through PCR (Polymerase Chain Reaction) identification, sequencing and virulence test, the virus is named as a PRV SD2020 strain, the strain is taken as a parent, a gI-gE gene and a TK gene of the strain are knocked out by utilizing a gene editing technology (CRISPR / CAS9 system), and a double-gene lost strain PRV SD2020 delta gI-gE and a three-gene lost strain PRV SD2020 delta gI-gE-TK are constructed. When the two gene deletion strains are used for preparing an inactivated vaccine and a live vaccine, an immune challenge protection test result shows that the prepared vaccine has good safety and immunogenicity, and shows that the two strains are suitable for preventing, controlling and purifying pseudorabies diseases.
Owner:JIANGXI ZHENGBANG TECHNOLOGY CO LTD
A method and application for detecting covid-19 based on mngs
ActiveCN113337639BImprove reverse transcription efficiencyLow pollution rateMicrobiological testing/measurementLibrary creationViral nucleic acidVariant virus
Owner:天津金匙医学科技有限公司
RNA vaccine for porcine epidemic diarrhea and construction method thereof
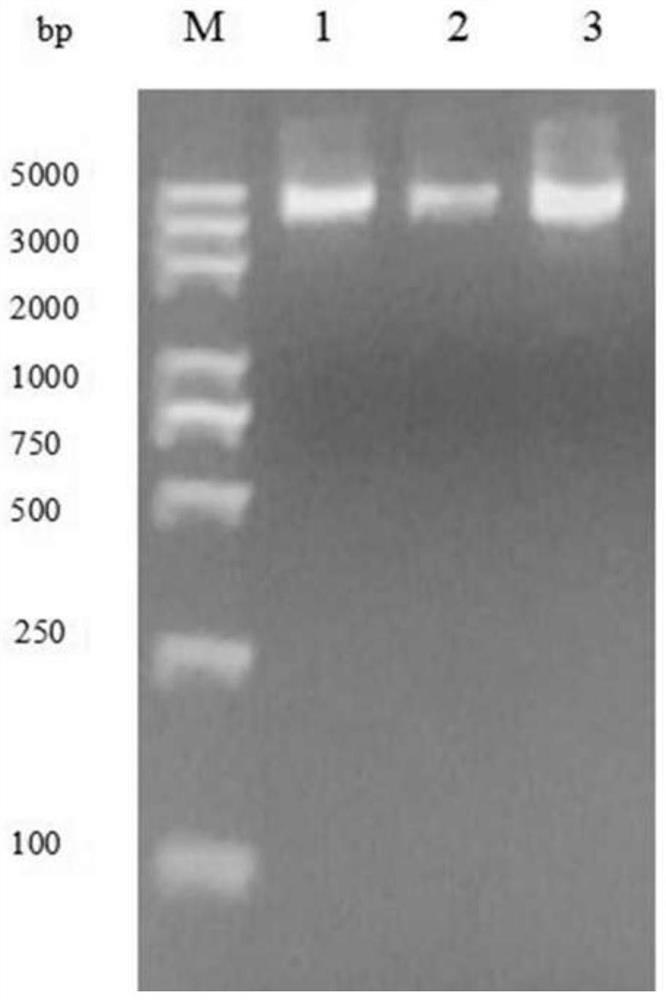
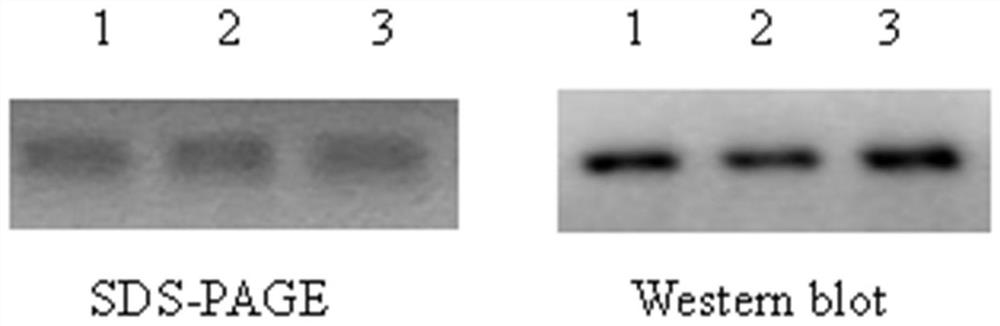
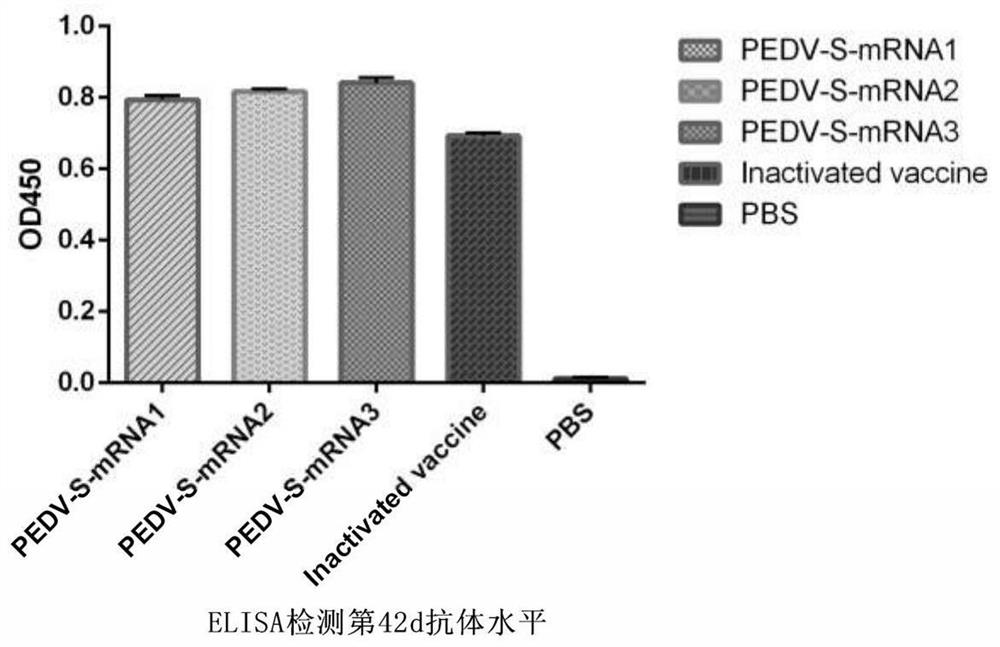
PendingCN113274491ALow costIncrease productivitySsRNA viruses positive-senseViral antigen ingredientsVariant virusTGE VACCINE
The invention belongs to the technical field of RNA vaccines, and particularly relates to an RNA vaccine for porcine epidemic diarrhea and a construction method thereof. According to construction of the RNA vaccine, porcine 5' UTR and 3' UTR are selected to be connected with a S protein, termination codon and polyA nucleotide sequence of a strain in series to obtain the RNA vaccine; by adopting a lipid nanoparticle as a delivery carrier of the RNA vaccine, a novel porcine epidemic diarrhea (PEDV) strain of a current epidemic wild strain can be rapidly coped, and the novel PEDV strain can be efficiently and rapidly targeted. The RNA vaccine for the porcine epidemic diarrhea, prepared by the invention, is low in cost and high in production efficiency, and can induce cellular immunity and humoral immunity at the same time; the prepared lipid nanoparticle-encapsulated RNA vaccine can rapidly cope the new PEDV strain of the current epidemic wild strain, especially a variant virulent strain, so that the research, development and production cycle of new vaccine development is shortened, and an effective technical means is provided for prevention and control of a new variant virulent strain.
Owner:广州源博医药科技有限公司
QRT-PCR (quantitative reverse transcription-polymerase chain reaction) method for identifying novel coronavirus Gamma variant
PendingCN114164303AReduce dependenceSave time for transshipmentMicrobiological testing/measurementAgainst vector-borne diseasesVariant virusTranscription (biology)
The invention belongs to the technical field of biology, and particularly relates to a qRT-PCR (quantitative reverse transcription-polymerase chain reaction) method for identifying a novel coronavirus Gamma variant. According to the method, in order to improve the detection sensitivity and specificity, a TaqMan probe is introduced; in order to reduce the cost, two pairs of primers are changed into one pair of half primers, that is, two upstream primers respectively target a mutation site and an original non-mutated site, and one downstream primer is shared by the two upstream primers. The sensitivity of variation detection is enhanced; mutation is introduced to the third site in the 5'direction of the mutation site of the upstream mutation primer, and by reducing the matching degree of the mutation primer and non-mutated virus nucleic acid and the matching degree of the non-mutated primer and the mutation virus nucleic acid in the reaction system, the amplification curve of the mutation primer for amplifying the mutation virus nucleic acid in the reaction system is earlier than the amplification curve of the non-mutated nucleic acid; the amplification curve of the non-variant nucleic acid amplified by the non-variant primer is earlier than the amplification curve of the variant nucleic acid amplified by the non-variant primer.
Owner:SHANXI UNIV
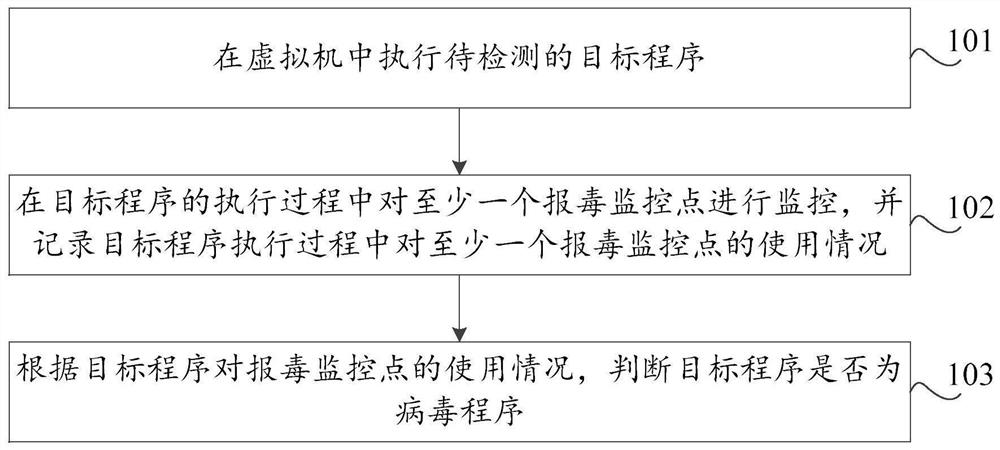
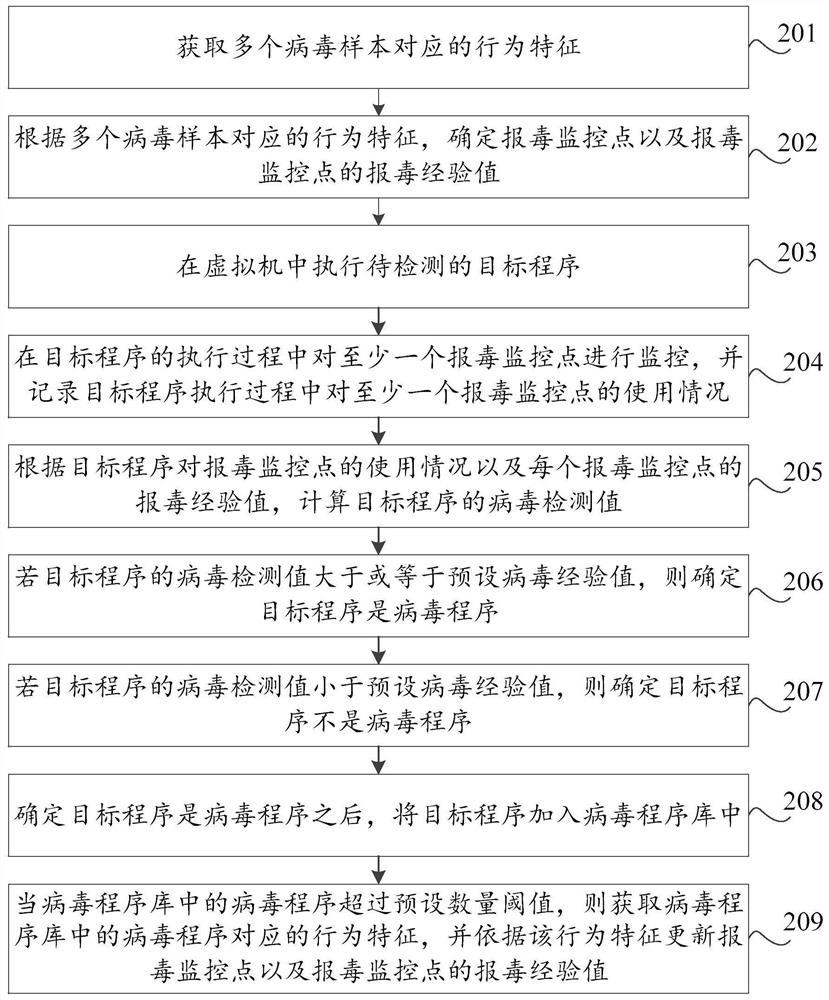
Virus reporting method and device based on virtual machine, storage medium and computer equipment
PendingCN112580025AProtection securityImprove detection ratePlatform integrity maintainanceVariant virusNovel virus
The invention discloses a poison reporting method and device based on a virtual machine, a storage medium and computer equipment. The poison reporting method comprises the steps of executing a to-be-detected target program in the virtual machine; monitoring at least one poison reporting monitoring point in the execution process of the target program, and recording the use condition of the at leastone poison reporting monitoring point in the execution process of the target program; and judging whether the target program is a virus program or not according to the use condition of the target program for the virus reporting monitoring point. By utilizing the virtual machine technology, the running record of the target program during execution in the virtual machine can be effectively obtained, the real computer environment is not influenced, the safety of the computer is protected, meanwhile, heuristic virus detection is carried out on the basis of the running record of the target program. The detection rate of unknown viruses such as novel viruses and variant viruses is favorably improved.
Owner:QI AN XIN SECURITY TECH ZHUHAI CO LTD +1
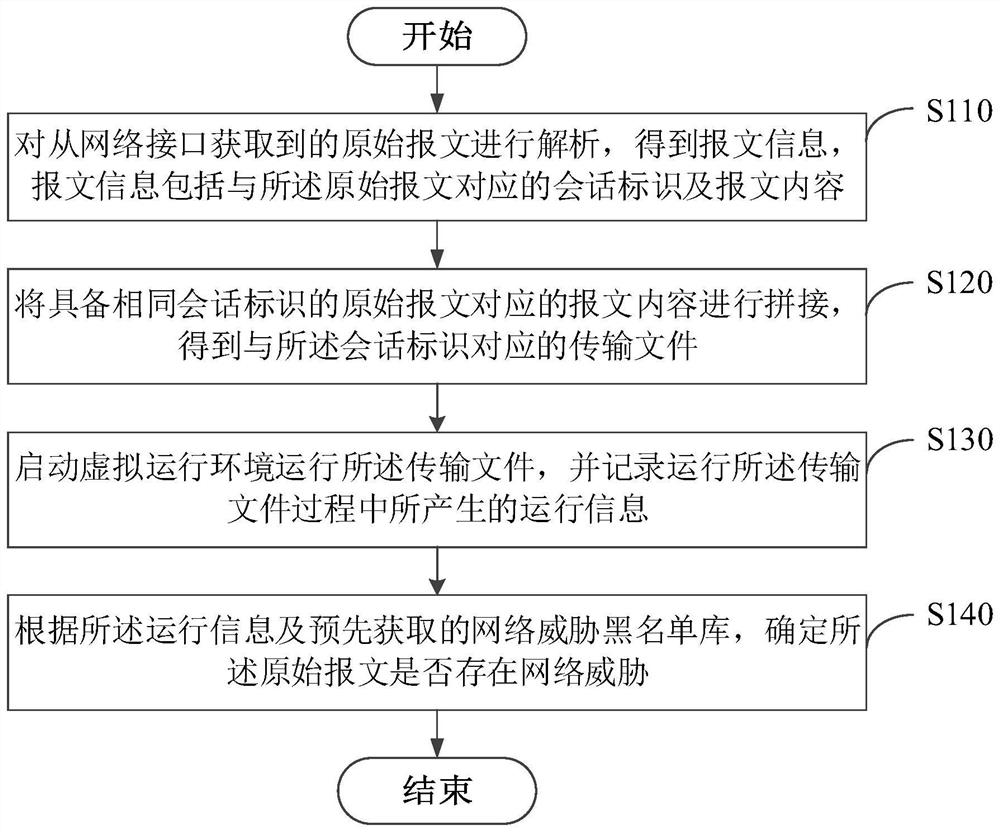
Network threat detection method, device and system, electronic equipment and storage medium
The invention relates to a network threat detection method, device and system, electronic equipment and a storage medium, and belongs to the field of network security. The method comprises the steps that when an original message is detected, the original message is restored into a transmission file, then a virtual running environment is started to run the transmission file, running information including intermediate behaviors triggered in the running process of the transmission file is obtained, and then by comparing the running information with a network threat blacklist library, whether the transmission file corresponding to the original message has the intermediate behavior causing the network threat in the operation process or not is determined, and whether the original message has the network threat or not is determined, so that the network threat caused by the fact that the dynamic flow cannot be detected or the flow of variant viruses exists in traditional static detection is avoided.
Owner:BEIJING TOPSEC NETWORK SECURITY TECH +2
Application of solidago decurrens in preparation of medicine for treating and preventing new coronavirus COVID-19 and variant virus thereof
The invention discloses an application of solidago decurrens in preparation of a medicine for treating and preventing a new coronavirus COVID-19 and a variant virus of the new coronavirus COVID-19, and a traditional Chinese medicine stock solution (substance) extracted from a natural plant solidago decurrens is proved to have a rapid, efficient and reliable effect of resisting the new coronavirus COVID-19 and the variant virus of the new coronavirus COVID-19 through clinical practice. An excipient, an auxiliary agent or a carrier is added into the plant extracting solution (substance) with the effect according to different requirements and a conventional method, and raw materials of different dosage forms are prepared. The compound can be applied to preparation of drugs for treating and preventing the new coronavirus COVID-19 and the variant virus thereof.
Owner:楼士荣
QRT-PCR (quantitative reverse transcription-polymerase chain reaction) method for identifying novel coronavirus Lambda variant
PendingCN114277194AReduce dependenceSave time for transshipmentMicrobiological testing/measurementMicroorganism based processesVariant virusMolecular biology
The invention belongs to the technical field of biology, and particularly relates to a qRT-PCR (quantitative reverse transcription-polymerase chain reaction) method for identifying a novel coronavirus Lambda variant. According to the method, in order to improve the detection sensitivity and specificity, a TaqMan probe is introduced; in order to reduce the cost, two pairs of primers are changed into one pair of half primers, that is, two upstream primers respectively target a mutation site and an original non-mutated site, and one downstream primer is shared by the two upstream primers. The sensitivity of variation detection is enhanced; mutation is introduced to the third site in the 5'direction of the mutation site of the upstream mutation primer, and by reducing the matching degree of the mutation primer and non-mutated virus nucleic acid and the matching degree of the non-mutated primer and the mutation virus nucleic acid in the reaction system, the amplification curve of the mutation primer for amplifying the mutation virus nucleic acid in the reaction system is earlier than the amplification curve of the non-mutated nucleic acid; the amplification curve of the non-variant nucleic acid amplified by the non-variant primer is earlier than the amplification curve of the variant nucleic acid amplified by the non-variant primer.
Owner:SHANXI UNIV
Porcine pseudorabies virus gI/gE double-gene-deleted strain and application thereof
PendingCN114045269AImprove securityImproving immunogenicityViral antigen ingredientsVirus peptidesDiseaseRabies
The invention belongs to the technical field of virology genetic engineering and preventive veterinary medicine, particularly relates to a gI / gE double-gene-deleted strain of a porcine pseudorabies epidemic variation virus, and further discloses a construction method of the gI / gE double-gene-deleted strain and application of the gene-deleted strain. A female parent of the porcine pseudorabies virus gene-deleted strain PRV-E6-delta gE / gI is a wild epidemic variant strain PRV-E6 independently separated from a disease material, and on the basis of the variant strain PRV-E6, a unique and simplified one-step double-knockout technology is adopted to knock out gI / gE double-virulence genes, so the porcine pseudorabies virus gene-deleted strain PRV-E6-gE / gI is obtained. The gB, gC and TK genes of the strain have the same evolutionary characteristics as prevalent variant virulent strains newly appearing after 2012, and have the advantages of high proliferation titer and strong virulence (wherein a median lethal dose to mice is 10<3.3> TCID50, and the morbidity of 2-to-3-weeks-old piglets is 10<5> TCID50).
Owner:BEIJING KEMUFENG BIOLOGICAL PHARMA +3
QRT-PCR (quantitative reverse transcription-polymerase chain reaction) method for identifying novel coronavirus Omicro variant BA-2 branch
PendingCN114657285AReduce dependenceSave time for transshipmentMicrobiological testing/measurementDNA/RNA fragmentationVariant virusTranscription (biology)
The invention belongs to the technical field of biology, and particularly relates to a qRT-PCR (quantitative reverse transcription-polymerase chain reaction) method for identifying a novel coronavirus Omicro variant BA-2 branch. According to the method, in order to improve the detection sensitivity and specificity, a TaqMan probe is introduced; in order to reduce the cost, two pairs of primers are changed into one pair of half primers, that is, two upstream primers respectively target a mutation site and an original non-mutated site, and one downstream primer is shared by the two upstream primers. By reducing the matching degree between the variation primer and the non-variation virus nucleic acid and the matching degree between the non-variation primer and the variation virus nucleic acid in the reaction system, the amplification curve of the variation primer for amplifying the variation virus nucleic acid in the reaction system is earlier than the amplification curve of the non-variation nucleic acid; the amplification curve of the non-variant nucleic acid amplified by the non-variant primer is earlier than the amplification curve of the variant nucleic acid amplified by the non-variant primer.
Owner:SHANXI UNIV
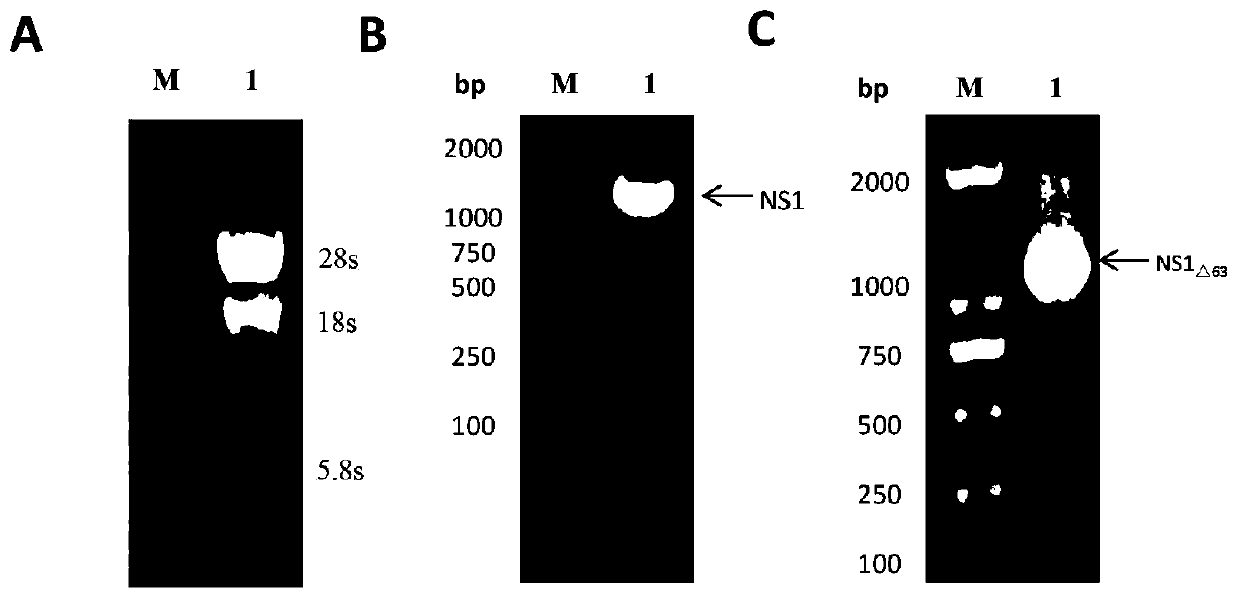
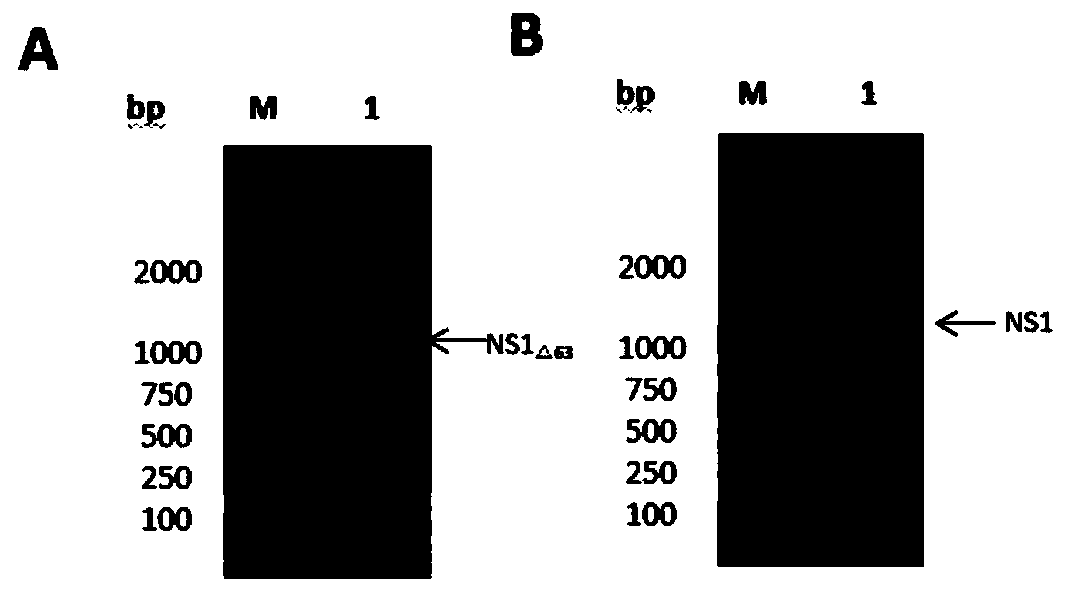
A Japanese encephalitis virus non-structural protein ns1 truncated mutant and its coding gene and application
ActiveCN108707191BEfficient and stable expressionSsRNA viruses positive-senseViral antigen ingredientsEscherichia coliViral Vaccine
The invention relates to a Japanese encephalitis virus non-structural protein NS1 truncated mutant, its coding gene and application, and belongs to the technical field of viruses. The non-structural protein NS1 truncation mutant of Japanese encephalitis virus deletes two strong hydrophobic regions at the C-terminus on the basis of the original NS1, and obtains a truncation mutant NS1 with 63 amino acids at the C-terminus deleted △63 Gene, cloned into the prokaryotic expression vector pET‑28a(+), and obtained the recombinant expression vector pET28a‑NS1 △63 , transformed into E. coli, induced by IPTG, NS1 △63 It can be expressed efficiently and stably in Escherichia coli. NS1 expressed in E. coli △63 After protein purification and immunization of mice, the titer of NS1 antiserum of immunized mice reached 1:12150 by ELISA. Protection tests in immunized mice showed that NS1 △63 Has protective activity. The NS1 △63 The protein can lay the foundation for the development of later virus vaccines and diagnostic kits.
Owner:JIANGXI AGRICULTURAL UNIVERSITY
Single clone antibody of antimutagen hepatitis B virus surface antigen and preparation method thereof
InactiveCN1284860CAvoid losing toQuality assuranceImmunoglobulins against virusesFused cellsMutation CarrierAntigen
A monoclonal antibody against mutant hepatitis B virus surface antigen secreted by hybridoma cell line CGMCC HBSP2. The anti-mutated hepatitis B surface antigen monoclonal antibody prepared according to the present invention can simultaneously detect hepatitis B viruses including 14 kinds of mutant strains and wild strains, and can replace the HBsAb in the conventional hepatitis B surface antigen detection kit. Into the hepatitis B surface antigen diagnostic kit. The use of the antibody can avoid the missed diagnosis caused by conventional reagents, so that the carriers of the mutated and non-mutated hepatitis B virus or hepatitis B patients can be diagnosed and treated in time. Especially when screening blood donors, it can detect mutant virus carriers that cannot be detected by conventional kits, avoid transfusing the blood of hepatitis B patients as healthy blood to recipients, and ensure the quality of blood sources.
Owner:徐钧
Anti-Trojan-horse-virus analyzer
InactiveCN104281807APrevent force closeLess time spent manually analyzing virusesPlatform integrity maintainanceVariant virusFalse alarm
The invention relates to an anti-Trojan-horse-virus analyzer which is developed based on the simulated anti-computer-virus engineer technology. Through the technology, feature codes can be fast analyzed and extracted through cooperation with an LDM threat analyzer, so that the cost for manually collecting the feature codes is greatly reduced, the time for manually collecting the feature codes is greatly shortened, and faster virus killing responses can be achieved. Variant viruses can be recognized with only one feature code, and the false alarm rate is extremely low.
Owner:SHANGHAI KEYU MARKETING PLANNING
Pure traditional Chinese medicine prescription composition used for purifying breeding sow disease-free pig farms, and avoiding death of sucking pigs with dysentery caused by virus mutants and viruses carried by breeding sows
InactiveCN107582882AImprove the problem that the sense of smell is very sensitive to bitter taste and not to eatRaise maternal antibodyAntibacterial agentsPowder deliveryBiotechnologyPig farms
The invention discloses a pure traditional Chinese medicine prescription composition used for purifying breeding sow disease-free pig farms, and avoiding death of sucking pigs with dysentery caused byvirus mutants and viruses carried by breeding sows. The pure traditional Chinese medicine prescription composition is composed of following raw materials via processing: radix bupleuri, dried FructusForsythiae, mulberry leaf, great burdock achene, reed rhizome, gentian, Flos Eriocauli, lophatherum gracile, cordate houttuynia, patrinia herb, pale butterfly bush flower, cassia seed, coptis chinensis, oriental waterplantain rhizome, Herba Artemisiae scopariae, rheum officinale, Ligusticum wallichii, motherwort, rhizoma corydalis, herba epimedii, sophora bud, Herba Agrimoniae, sanguisorba officinalis, artificially cultured Radix Scutellariae, artificially cultured Radix Astragali, bighead atractylodes rhizome, magnolia cortex, and licorice. According to a preparation method, the above 28 traditional Chinese medicinal materials are prepared into powder via different processing methods (stir-baking with vinegar, wine, or ginger juice), the pure traditional Chinese medicine prescription composition is capable of solving a problem that pure traditional Chinese medicines are not accepted by breeding sows because of the obvious bitter taste, and avoiding death of sucking pigs with dysentery caused by virus mutants and viruses carried by breeding sows, and is safe and reliable; no toxic or side effect is caused; and no drug residue is left.
Owner:河南省兽用纯中药研究院有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com