Method for detecting COVID-19 based on mNGS and application of method
A COVID-19, multi-purpose technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, resistance to vector-borne diseases, etc., can solve problems such as false negatives, reduce the enrichment effect of mutant virus sequences, and achieve growth and extension length, improve reverse transcription efficiency, and ensure high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
experiment example 1
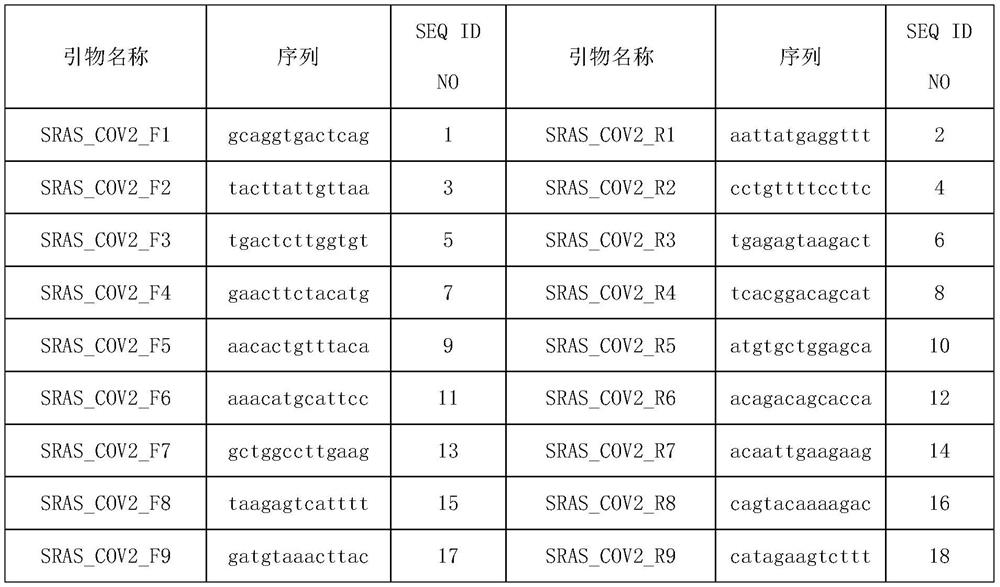
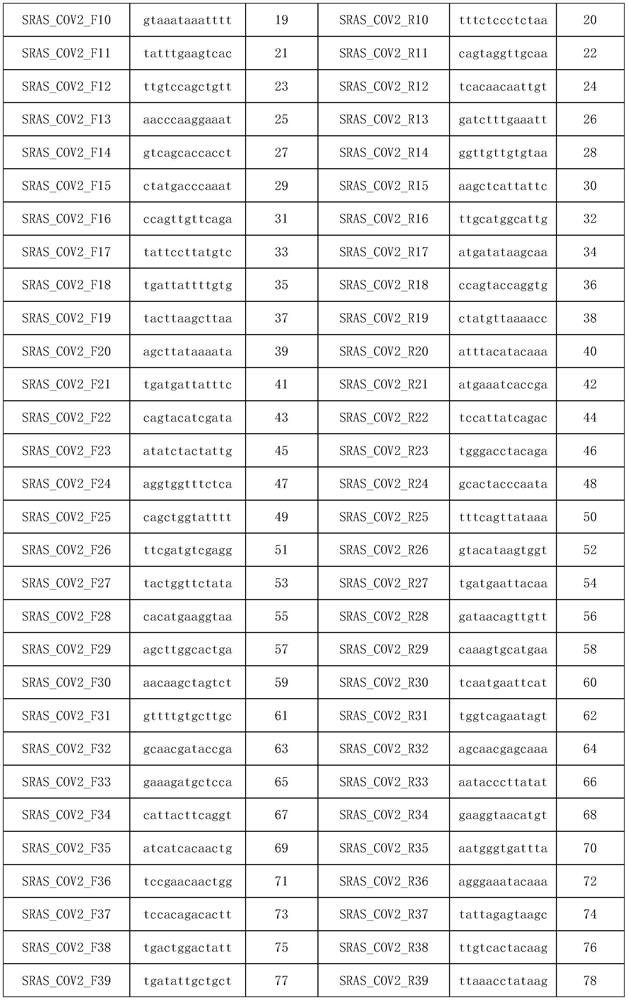
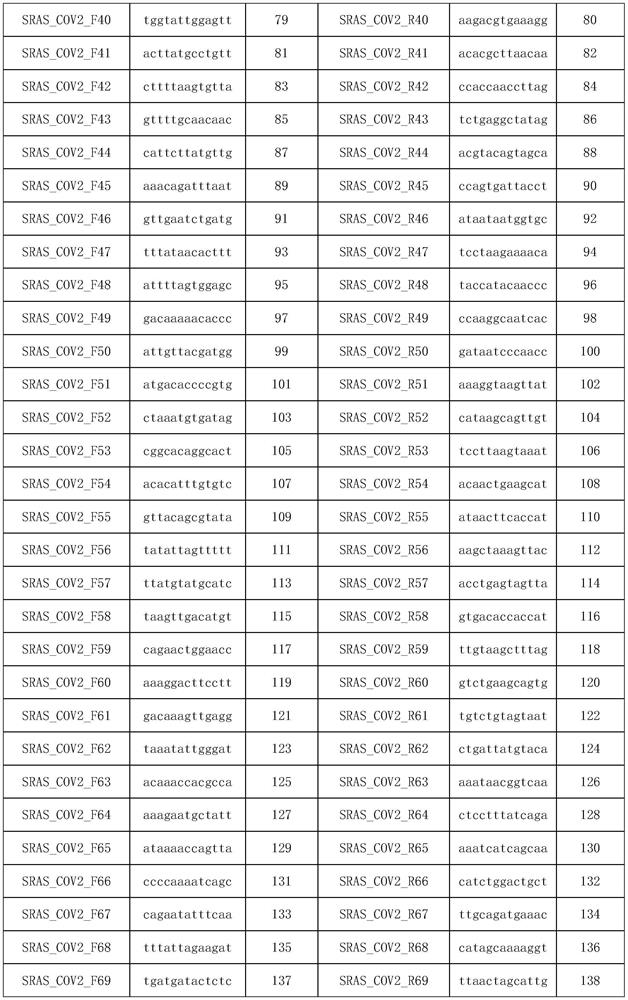
[0066] Experimental Example 1 Primer Design
[0067] 1. Generate multiple genomes
[0068] First download more than 100 COVID-19 novel coronavirus genomes from the NCBI website, and use fast Fourier transform (MAFFT) (v.7.388) to match and align all the downloaded genomes; Note, to ensure the mutation detection rate of subsequent primers , where the source sequences of multiple genomes must be at least greater than 100.
[0069] 2. Candidate primer design.
[0070] Use PYFASTA software to segment the matched multiple genomes into 394nt short fragments with 197nt overlap, and intercept 50nt at both ends of each fragment as the primer design region, design multiple 13nt forward or reverse primers to form the region The primer set to be merged, and then all the primers in the primer set to be merged in each region are sorted from high to low frequency, select the primer with the highest frequency and does not contain uncertain bases, and then delete the primer sequence All the...
Embodiment 1
[0120] Example 1 Experimental verification of reverse transcription primer set
[0121] 1. Sample source
[0122] The new coronavirus pseudovirus used in the preparation of the positive reference product was purchased from Shanghai Yisheng, using a retroviral vector loaded with the partial sequence of the COVID-19 virus ORF1 a / b gene, the entire sequence of the E gene and the N gene coding region, with a length of 2000bp . It was verified positive by in vitro diagnostic reagents (fluorescent quantitative PCR method), and the standard curve was quantified as 2*10^8copies / mL. The cells used were purchased from Nanjing Kebai, and the cell pellet was 10^6 / tube.
[0123] 2. Experimental sample preparation
[0124] Since the detection limit of metagenomic detection of RNA viruses in clinical respiratory samples is expected to be 1000 copies / mL, the concentration of COVID-19 for preparing positive samples is 1×10^3 copies / mL. In order to simulate the host content of alveolar lavag...
Embodiment 2
[0136] Example 2: Experimental verification of the reverse transcription primer set (the effect of the primer set on detection enhancement).
[0137] 1. The sample source is the same as in Example 1.
[0138] 2. Experimental sample preparation
[0139] Prepare positive samples P1 and P2 with concentrations of 1×10^3copies / mL and 1×10^4copies / mL of COVID-19 pseudovirus respectively. In order to simulate the host content of alveolar lavage fluid, cultured human cell lines were added to P1 and P2 to make the cell concentration 10^5Cell / mL. And prepare a negative sample N1 without pseudovirus.
[0140] 3. Experimental method
[0141] The experimental scheme selected in Example 1 was adopted, that is, after ribosomal RNA was removed, full-length reverse transcription was performed using specific primers and random primers, and then the double-stranded cDNA was digested and interrupted to build a library, compared with and without COVID-19 The number of reads detected by multipl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com