A polymerase mutant and its application
A mutant and polymerase technology, applied in the field of molecular biology, can solve the problems of DNA chain duplication of double-stranded DNA, inability to use catalytic loop to mediate isothermal amplification, etc., to achieve increased specificity, high cDNA synthesis efficiency and detection sensitivity , the effect of improving enzyme concentration
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Expression and purification of polymerase mutants
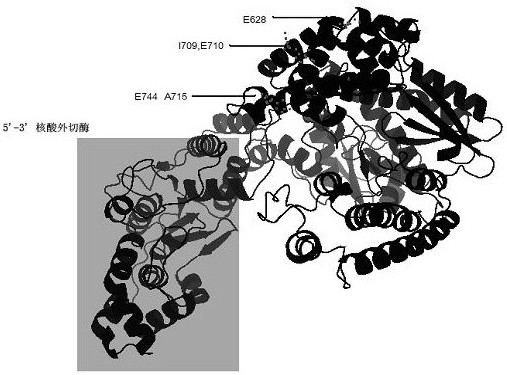
[0034] 1. Preparation of polymerase mutant gene expression vector. Take the plasmid template of ZO5 wild-type polymerase gene (SEQ ID NO: 2), and use E628K-F, E628K-R, IE709LK-F, IE709LK-R, EA744RR-F, EA744RR-R as mutation primers, and use ZO5- Nde1-F and ZO5-Sal1-R were truncated PCR primers, and 1 μL of primers (10 μM) were added to a 50 μL reaction system. The mix uses a PCR reaction system of 50 μL, including 10 × buffer (100 mM KCl, 100 mM (NH 4 ) 2 SO 4 , 200mM Tris-HCl pH=8.8, 20mM MgSO 4, 5 μL of 1% TritonX-100, 1 mg / mL BSA), 3 μL of 2 mM dNTPs, 1 μL of KOD enzyme. A total of 25 cycles of amplification program: 95°C, 1min; 95°C, 30s; 60°C, 30s; 68°C, 2min; 68°C, 5min, 4°C preservation. Amplification primers are shown in Table 1. Among them, IE709LK stands for I709L, E710K, EA744RR stands for E744R, A745R.
[0035] Table 1 Amplification primer sequences
[0036] primer name sequen...
Embodiment 2
[0039] Example 2: RT-PCR of polymerase mutants
[0040] The RNA prepared by the DNA sequence of MS2 of the published patent CN113846146A was used as a template (DNA sequence shown in SEQ ID NO: 8), and each well contained 3 μL of ZO5 L1 and ZO5 L2 enzymes diluted 10 times with buffer, ZO5 wild-type polymerase was used as a control for PCR and RT-PCR. In each group, the buffer contained 20 mM Tris-HCl (pH=8), 100 mM KCl, 0.1 mM EDTA, 0.1% Tween-2. Each was added to 12 μL of RT-PCR master mix as shown in Table 2, and thermocycling was performed. Thermal cycling conditions were: 50°C, 2 minutes ("UNG" step); 65°C, 2 minutes ("RT" step); 5 cycles of 94°C, 15 seconds; followed by 62°C, 30 seconds; One cycle of 91°C, 15 seconds; followed by 62°C, 30 seconds.
[0041] Table 2 RT-PCR master mix
[0042] component concentration Tris-HCl (pH=8) 50mM KOAc 60mM glycerin 5% (v / v) DMSO 2% (v / v) MgCl2 2mM Primer 1 200nM Primer 1 ...
Embodiment 3
[0044] Example 3: Reaction temperature test of LAMP of polymerase mutants
[0045] Design primers to detect MS2-specific genes (MS2 gene sequence is shown in SEQ ID NO: 8), the design of MS2 primer sequences is shown in Table 3, and the reaction system is shown in Table 4.
[0046] Table 3 MS2 primer sequences
[0047] primer name sequence (5'to3') MS2-BIP GCACGTTCTCCAACGGTGCTGGTTGCTTGTTCAGCGAACT MS2-FIP GCCCAAACAACGACGATCGGTAAAACCAGCATCCGTAGCCT MS2-LB TGCAGGATGCAGCGCCTTA MS2-LF CCAGAGAGGAGGTTGCCAA MS2-B3 CAATAGAGCCGCTCTCAGAG MS2-F3 TGTCATGGGATCCGGATGTT
[0048] Table 4 Reaction system
[0049] sample concentration Addition amount (uL) MS2 Template 1ng / uL 1 MS2-BIP 10um 1.6 MS2-FIP 10um 1.6 MS2-LF 10um 0.8 MS2-LB 10um 0.8 MS2-F3 10um 0.2 MS2-B3 10um 0.2 dNTP / dUTP Mixture 25mM 0.8 Mg 2+
100mM 1 10×buffer \ 2.5 DNA polym...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com