A SNP molecular marker related to hypoxia tolerance traits of pomfret ovata and its application
A technology of oval pomfret and molecular marker, which can be applied in recombinant DNA technology, microbial determination/test, DNA/RNA fragment, etc., can solve the problem that the research on the marker of oval pomfret tolerance to hypoxia has not been reported, etc. Achieve the effect of improving genetic level and enriching resources
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
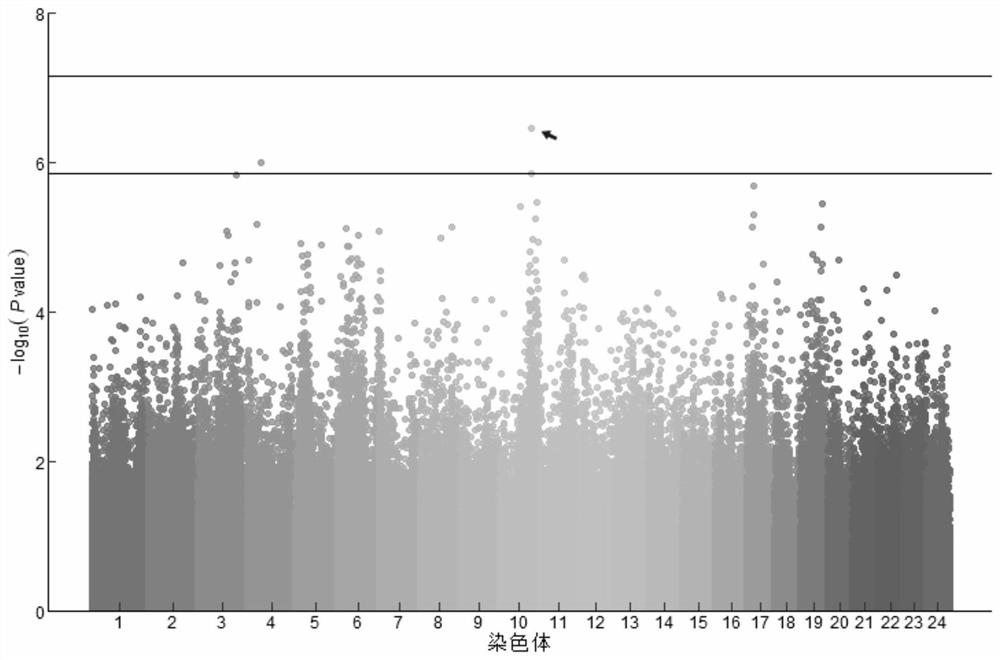
[0062] Screening of SNP markers:
[0063] (1) Randomly select 500 pompano pomfrets that have been treated with dissolved oxygen in water below the suffocation point, and record the death time of each fish as the basis for judging its hypoxia tolerance;
[0064] (2) Cut out the fin rays of 50 ovate pomfrets that died first and last, and extract DNA using the magnetic bead method DNA extraction kit, take 20 mg of fin rays, and add reagents to extract ovate pompano according to the kit instructions Trevally DNA. The extracted DNA was stored in 150 μL Buffer AE, and the DNA sample was detected by 1% agarose gel electrophoresis, and the DNA concentration and purity were detected by Nanodrop2000. The ratio of A260 / A280 was required to be between 1.8 and 2.0, and the DNA concentration was greater than 100 μg / μL.
[0065] (3) Genomic DNA was randomly broken into 350bp fragments, after end repair, phosphorylation, and A-tailing, the two ends of the fragments were respectively connecte...
Embodiment 2
[0075] Detection of SNP genotype:
[0076] The above-mentioned SNP molecular markers of the present invention were verified in another germplasm population containing 497 ovate pomfrets.
[0077] To determine the hypoxic tolerance of pomfret ovata, that is, the survival time under dissolved oxygen lower than the suffocation point. Concrete measuring method is with embodiment 1.
[0078] (1) Cutting part of the fin rays of the oval pomfret to extract the total DNA, the extraction and storage method and DNA quality requirements are the same as those described in Example 1. Use SNP primer software to design specific amplification primers based on the base sequence of 120 bp upstream and downstream of the SNP marker described in the present invention as a template.
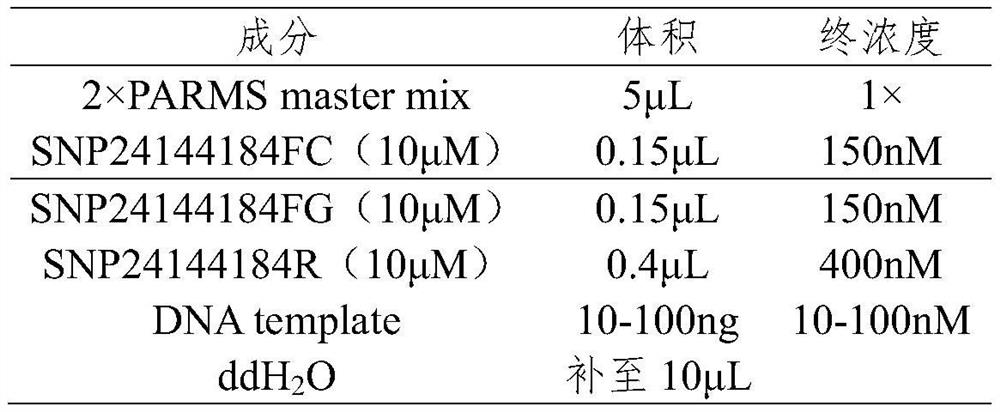
[0079] (2) PCR amplification
[0080] PCR primers:
[0081] SNP24144184FC (SEQ ID NO. 2):
[0082] GAAGGTGACCAAGTTCATGCTGGCTGAAGTCAGAGATGTGACTCTC
[0083] SNP24144184FG (SEQ ID NO. 3):
[0084] GAAGGTCGGAGTCAAC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com