Microbial metagenome binning method and system
A metagenomic and microorganism technology, applied in the field of microbial metagenomic binning methods and systems, can solve problems such as loss of useful features, low binning accuracy, and inability to accurately describe sequence features of microorganisms, so as to achieve accurate binning results and improve processing. Efficiency, the effect of reducing feature dimension
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] The purpose of this embodiment is to provide a microbial metagenomic composition box method.
[0034] A microbial metagenomic component box method, comprising:
[0035] Obtain the microbial metagenomic sequence to be binned;
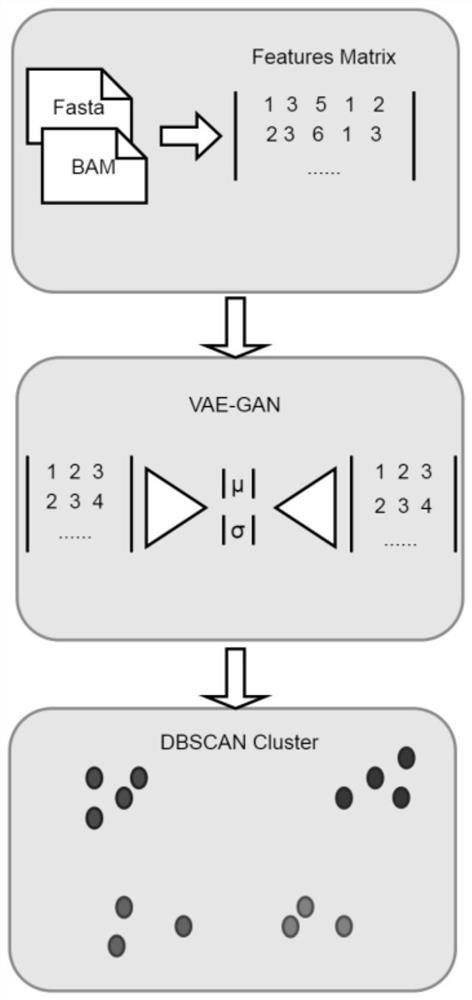
[0036] Feature extraction is performed on each sequence in the metagenomic sequence, wherein the extracted features include tetranucleotide frequency, RPKM abundance and kmer coverage features;
[0037] Input the extracted features into the VAE-GAN neural network for training, and encode the extracted features into the VAE hidden vector through training;
[0038] Based on the mean variable in the VAE hidden vector, the metagenomic sequence is clustered, and the binning of the metagenomic set is realized according to the clustering result.
[0039] Specifically, for ease of understanding, the solutions described in the present disclosure will be described in detail below in conjunction with the accompanying drawings:
[0040] Such as figure 1 T...
Embodiment 2
[0066] The purpose of this embodiment is a microbial metagenomic component box system, comprising:
[0067] A data acquisition unit, which is used to acquire microbial metagenomic sequences to be binned;
[0068] A feature extraction unit, which is used to extract features from each sequence in the metagenomic sequence, wherein the extracted features include tetranucleotide frequency, RPKM abundance and kmer coverage features;
[0069] A feature dimension reduction unit, which is used to input the extracted features into the VAE-GAN neural network for training, and encode the extracted features into the VAE hidden vector through training;
[0070] A clustering unit is configured to cluster the metagenomic sequences based on the mean variable in the VAE hidden vector, and implement binning of the metagenomics according to the clustering result.
[0071] In further embodiments, there is also provided:
[0072] An electronic device includes a memory, a processor, and computer i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com