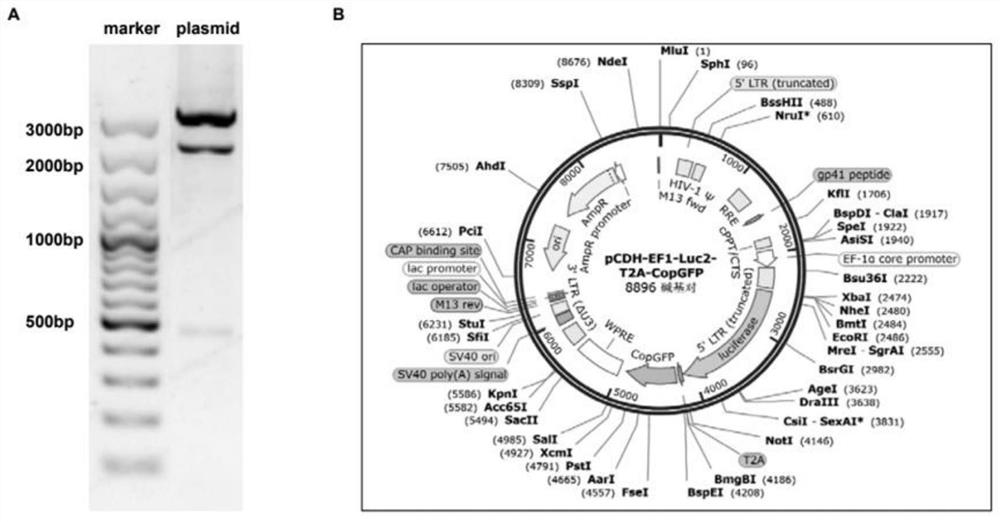
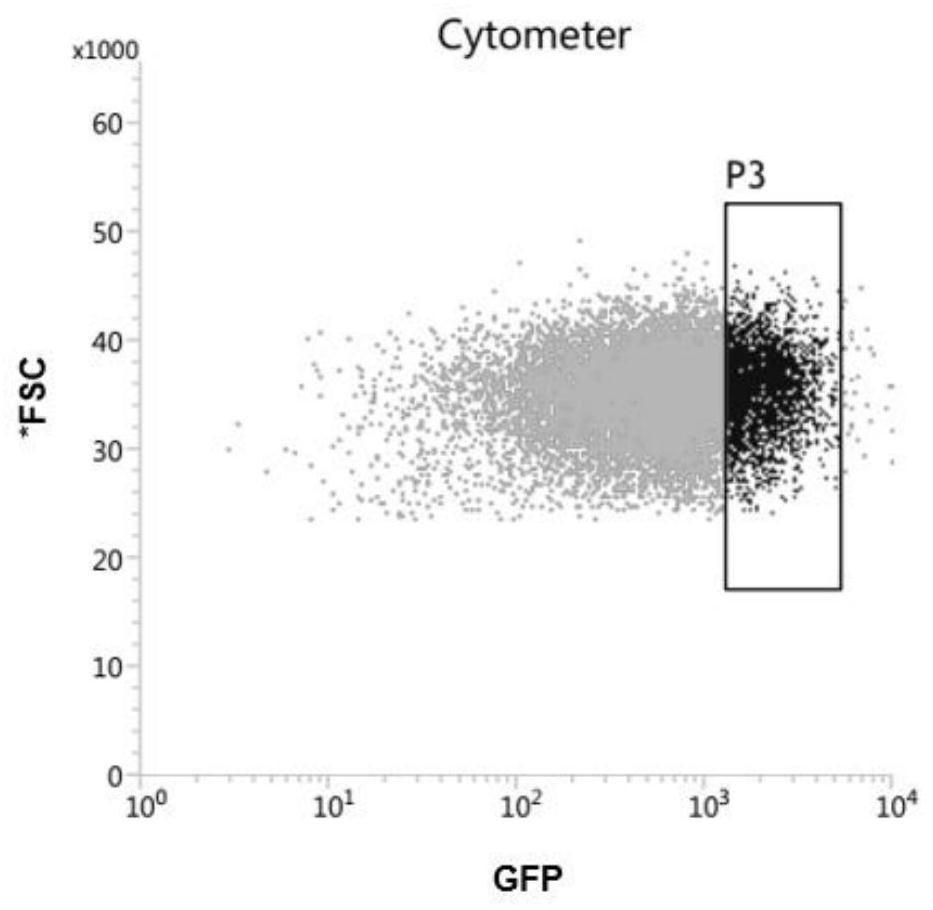
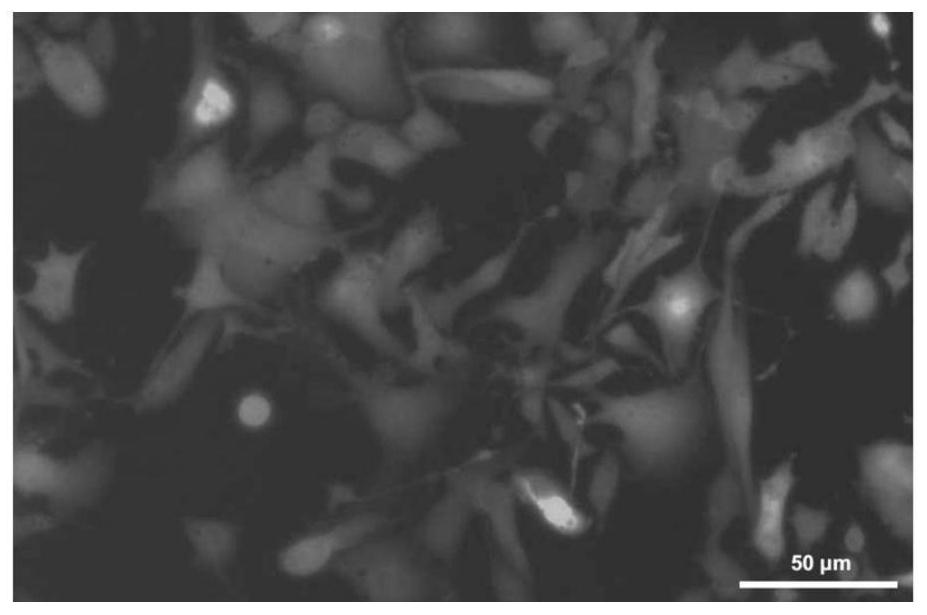
Melanoma leptomeningeal metastasis model construction method
A technology of metastasis model and construction method, applied in the biological field, can solve the problem of lack of animal models of melanoma leptomeningeal metastasis, and achieve the effects of convenient cell analysis, sorting and observation, high success rate and stability, and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] step 1:
[0032] In a cell culture dish with a diameter of 10 cm, the B16 melanoma cell line was cultured in RPMI-1640 complete medium (containing 10% FBS and 1% penicillin-streptomycin solution). In a cell culture dish with a diameter of 10 cm, 293T cell cultures were cultivated in DMEM complete medium (containing 10% FBS and 1% penicillin-streptomycin solution). The temperature of the cell culture incubator was 37 °C, CO 2 The concentration is 5%.
[0033] Step 2:
[0034] Using the pCDH-EF1-Luc2-T2A-Neo plasmid (concentration 100ng / μL) as a template, the Luciferase fragment was amplified by polymerase chain reaction (PCR) with the designed forward / reverse primers.
[0035] The base sequence of the forward primer is: 5'- CCTACTCTAGAGCTAGCGAATTCATGGAAG -3';
[0036] The base sequence of the reverse primer is: 5'- GAAGACTTCCTCTGCCCTCAG -3';
[0037] The PCR program used to amplify the Luciferase fragment is: 94°C pre-denaturation, 2 minutes; 94°C denaturation, 15 s...
Embodiment 2
[0051] In order to test the pial transfer ability of the screened B16-Luc2-CopGFP LM cells, we divided 5×10^ 4 Two B16-Luc2-CopGFPPar and B16-Luc2-CopGFP LM cells were injected into the left ventricle of mice, and the ability of the two kinds of cells to transfer to pia mater was observed and compared. Such as Figure 6 As shown, 15 days after cardiac injection, B16-Luc2-CopGFP LM cells diffused in large numbers in the pial space ( Figure 6 A), The screened B16-Luc2-CopGFP LM cells have a strong ability to metastasize to the pia mater. And all mice injected with B16-Luc2-CopGFP LM cells died within 20 days after cardiac injection ( Figure 6 B).
Embodiment 3
[0053] In order to further test the pial colonization ability of the screened B16-Luc2-CopGFP LM cells, we divided 2×10 4 B16-Luc2-CopGFPPar and B16-Luc2-CopGFP LM cells were injected into the cisterna magna of mice, and the survival of mice was observed and compared. Such as Figure 7 As shown, mice injected with B16-Luc2-CopGFP LM cells began to die 10 days after injection, and all died within two weeks, while no death was observed in mice injected with B16-Luc2-CopGFP Par cells. This indicated that the B16-Luc2-CopGFP LM cells screened for directional pial transfer had a high colonization ability in the pial space.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com