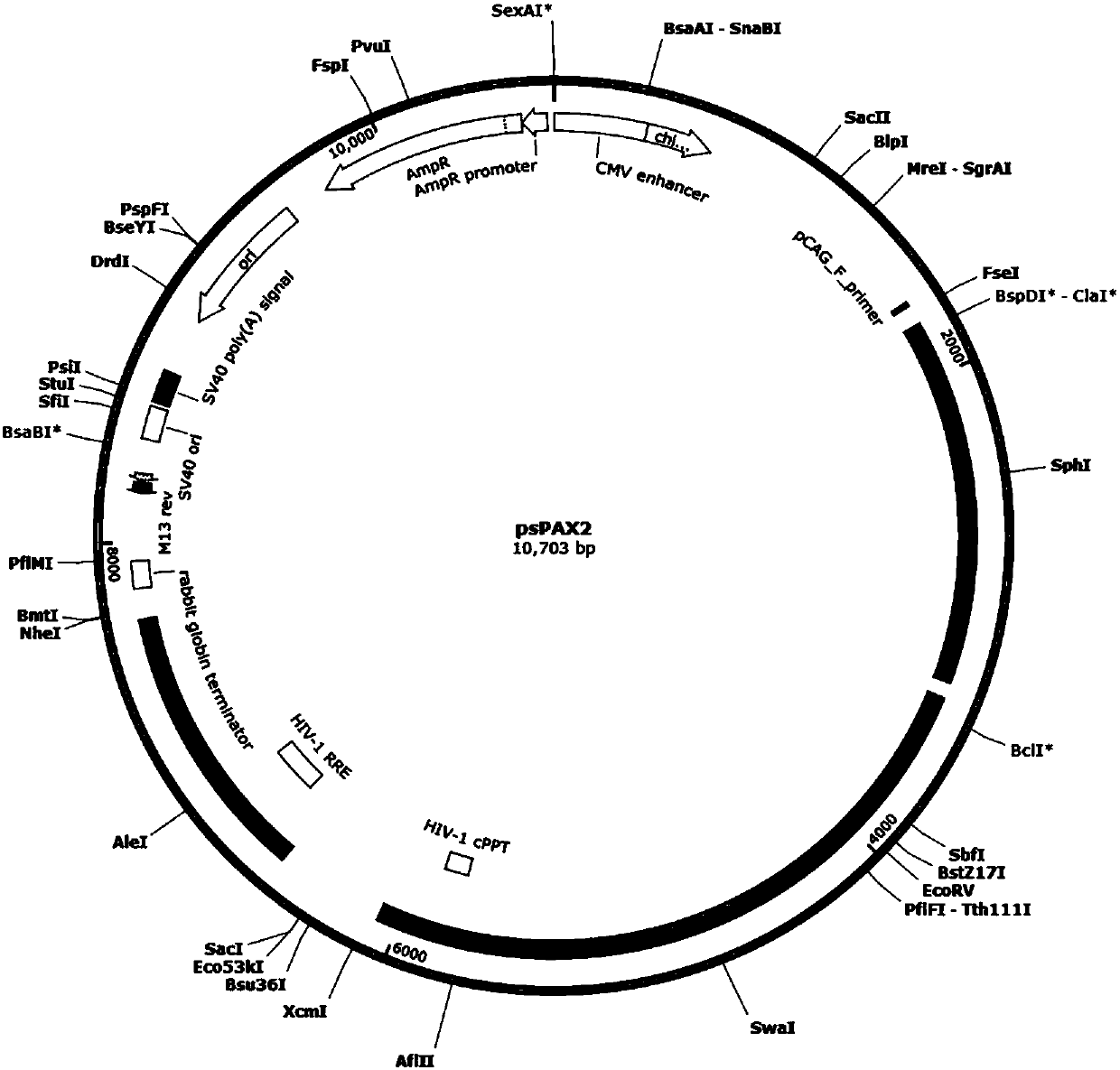
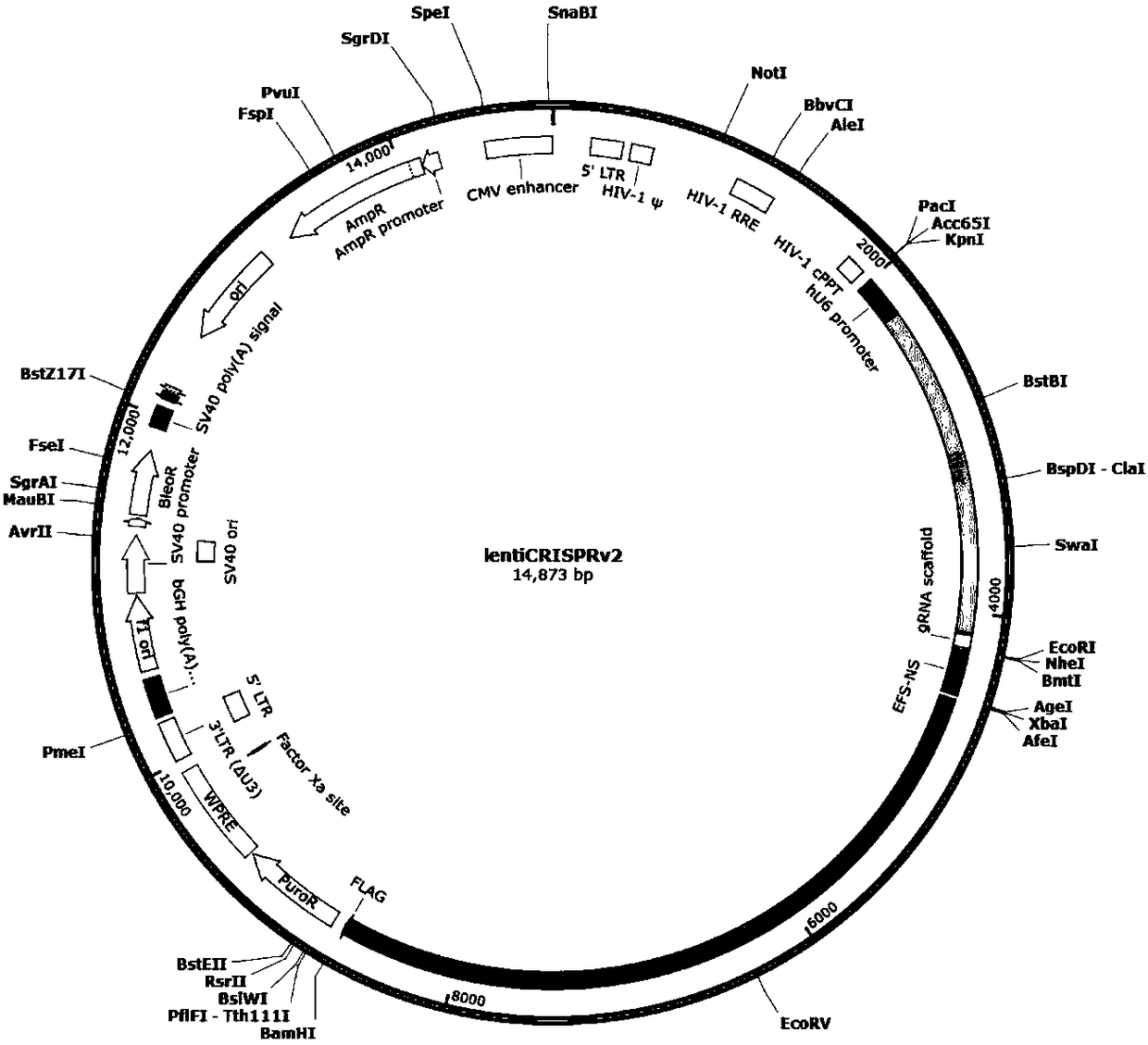
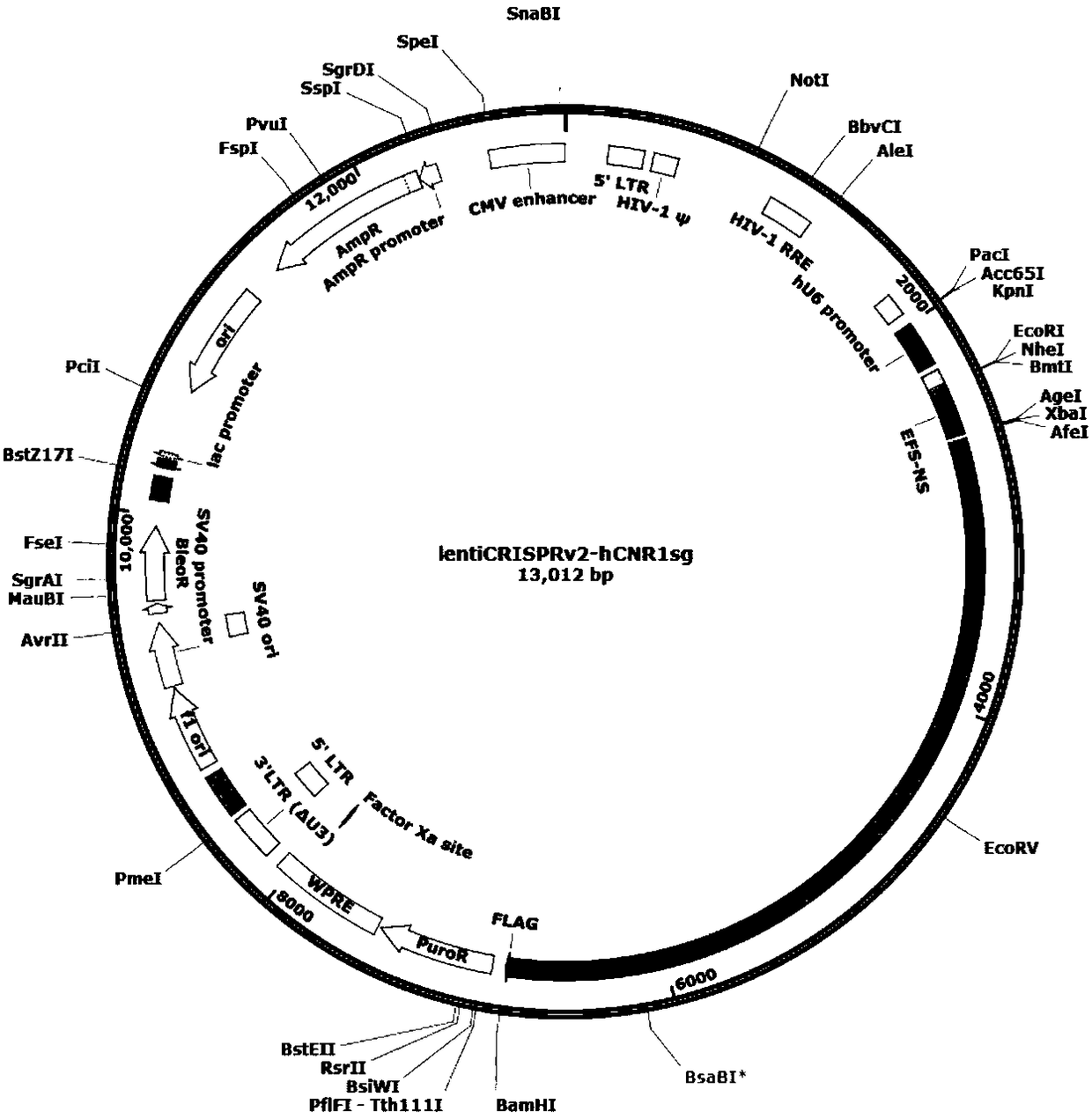
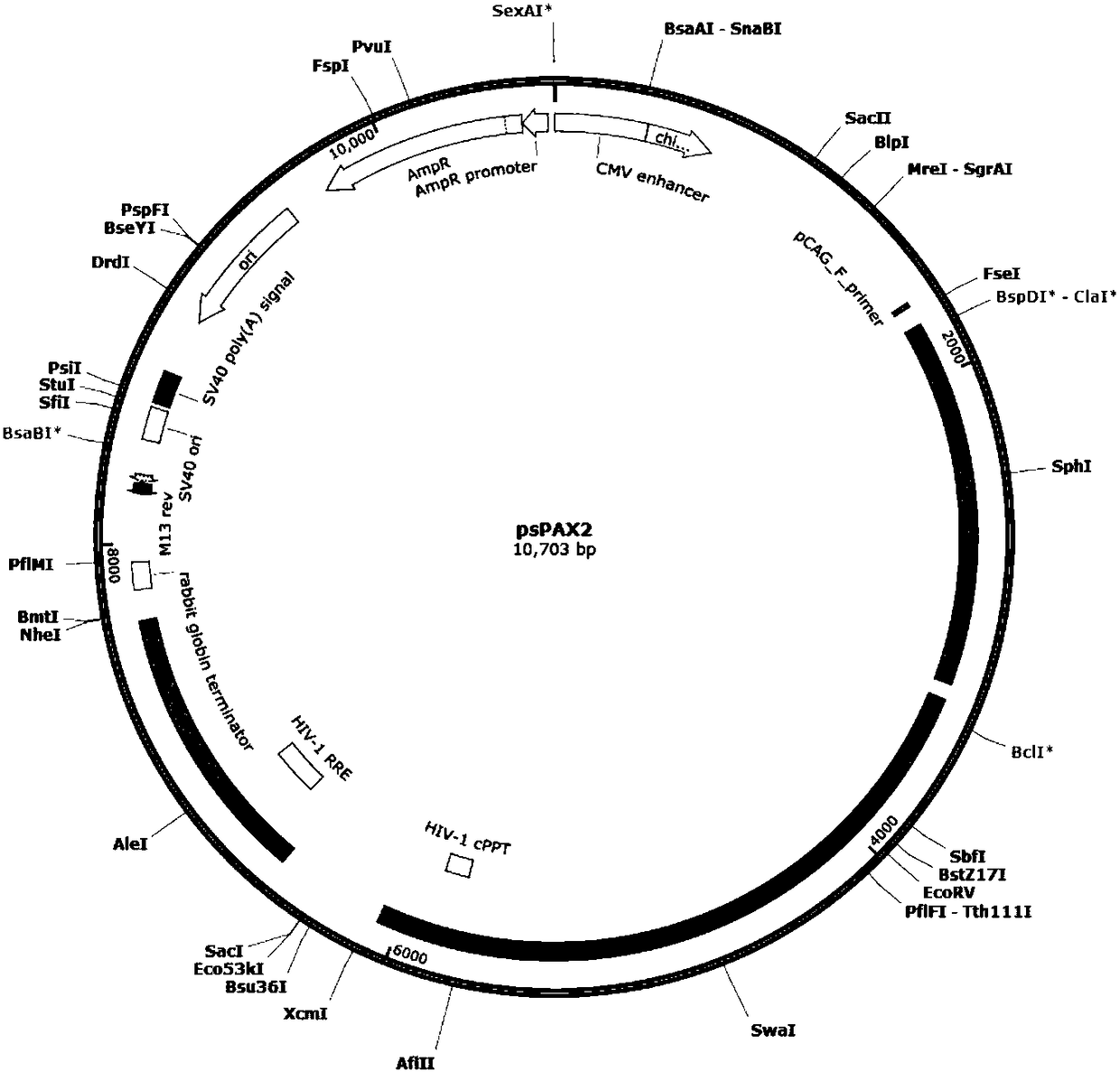
Patents
Literature
113 results about "Lentivirus Infections" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Virus diseases caused by the Lentivirus genus. They are multi-organ diseases characterized by long incubation periods and persistent infection.
Process of knocking out Wnt3a gene and verification method thereof
InactiveCN106434752AHigh targeting accuracyShort experiment cycleFermentationVector-based foreign material introductionValidation methodsWNT3A gene
The invention discloses a process of knocking out Wnt3a gene and a verification method thereof. The knockout and verification of Wnt3a gene are finished through the following steps: establishment of a Cas9 lentiviral vector for Wnt3a gene, culture and passage of HepG2 cell, lentivirus infection and screening of target cell, verification of gene knockout efficiency through a mispairing enzyme method, cell protein analysis and cell proliferation detection by a CCK-8 method. The invention has the following advantages: the Wnt3a gene is knocked out by establishing a Cas9 double-vector lentivirus system for the first time; Crispr / Cas9 is a technology for accurately editing specific site of the genome of any species, and the cell-level single gene or multiple genes can be knocked out by the technology; compared with other gene editing technologies, the method has the advantages that the targeting accuracy is higher; only if the RNA target sequence is completely matched with the genome sequence, can the Cas9 cut the DNA and realize simultaneous knockout of multiple sites of the target gene; and moreover, the experimental period of vector establishment is short, the time and the cost are remarkably saved, and species limit is avoided.
Owner:AFFILIATED HOSPITAL OF NANTONG UNIV
CRISPR/Cas9 targeted knockout human intestinal cancer cell RITA gene and specific sgRNA thereof
InactiveCN107893075AConvenient researchGenetically modified cellsNucleic acid vectorIntestinal CancerLentivirus
The present invention discloses CRISPR / Cas9 targeted knockout human intestinal cancer cell RITA gene and specific sgRNA thereof. First, sgRNA specifically targeted to a second exon of the RITA gene isobtained, and the base sequence of the sgRNA is as shown in SEQ ID NO. 1; secondly, a sgRNA lentiviral vector system of the RITA gene is constructed, and the sgRNA lentiviral vector system contains Cas9 protein; and finally human intestinal cancer HT-29 cells are infected with CRISPR / Cas9 lentivirus containing the sgRNA to obtain a cell line which is significantly reduced in RITA protein expression level. The invention has the advantages of simple operation steps, good sgRNA targeting property, and high RITA gene cutting efficiency; furthermore, the constructed CRISPR / Cas9 lentivirus system has the advantage of high knock-out efficiency and can specifically knock out the RITA gene to obtain RITA gene-knockout human intestinal cancer cells, and a powerful tool for the further study of theaction mechanism of RITA in the intestinal cancer cells is provided.
Owner:OBIO TECH SHANGHAI CORP LTD
CRISPR-Cas9 for targeting knockout of human intestinal cancer cell CNR1 gene, and specific sgRNA thereof
InactiveCN108588071AFacilitate the study of the mechanism of actionStable introduction of DNAFermentationIntestinal CancerLentivirus
The invention discloses a CRISPR-Cas9 for targeting knockout of a human intestinal cancer cell CNR1 gene, and specific sgRNA sequences thereof. Firstly, an sgRNA specifically targeting CNR1 gene second exon is obtained, and base sequences of the sgRNA are as shown in SEQ ID NO. 1; secondly, the sgRNA of the CNR1 gene is constructed to a lentiviral vector system, and the lentiviral vector system contains Cas9 protein; and finally, a CRISPR / Cas9 lentivirus containing the sgRNA is infected with human intestinal cancer HT-29 cells, and a cell strain with CNR1 protein expression level significantlybeing reduced is obtained. The operation steps are simple, good sgRNA targeting is achieved, and the cutting efficiency to the CNR1 gene is high; and in addition, the constructed CRISPR / Cas9 lentiviral system has the advantage of high knockout efficiency, and can specifically knock out the CNR1 gene, human intestinal cancer cells with the CNR1 gene being knocked out is obtained, and therefore a powerful tool for further study of the mechanism of action of CNR1 in intestinal cancer cells is provided.
Owner:OBIO TECH SHANGHAI CORP LTD
CRISPR-Cas9 targeted knockout human colorectal carcinoma cell DEAF1 gene and specific sgRNA thereof
InactiveCN108396027AConvenient researchGenetically modified cellsStable introduction of DNAVector systemLentivirus
The invention discloses a CRISPR / Cas9 targeted knockout human colorectal carcinoma cell DEAF1 gene and specific sgRNA thereof. The CRISPR / Cas9 targeted knockout human colorectal carcinoma cell DEAF1 gene and the specific sgRNA thereof are characterized in that: firstly, sgRNA of a second exon of the specific targeted DEAF1 gene is obtained and the base sequence of the sgRNA is shown as SEQ ID NO.1; secondly, the sgRNA of the DEAF1 gene is constructed into a lentiviral vector system, which contains Cas9 protein; finally, the CRISPR / Cas9 lentivirus containing the sgRNA is infected with human colorectal carcinoma cell HT-29 cell, so that a cell strain of which DEAF1 protein expression level is obviously reduced is obtained. The CRISPR / Cas9 targeted knockout human colorectal carcinoma cell DEAF1 gene disclosed by the invention has the advantages of simple operation steps, good sgRNA target ability and high cutting efficiency for the DEAF1 gene; in addition, the constructed CRISPR / Cas9 lentiviral vector system has the advantage of high knockout efficiency and can specifically knock out the DEAF1 gene to obtain the human colorectal carcinoma cells knocking out the DEAF1 gene, and therebya powerful tool is provided for further studying an action mechanism of DEAF1 in the colorectal carcinoma cells.
Owner:OBIO TECH SHANGHAI CORP LTD
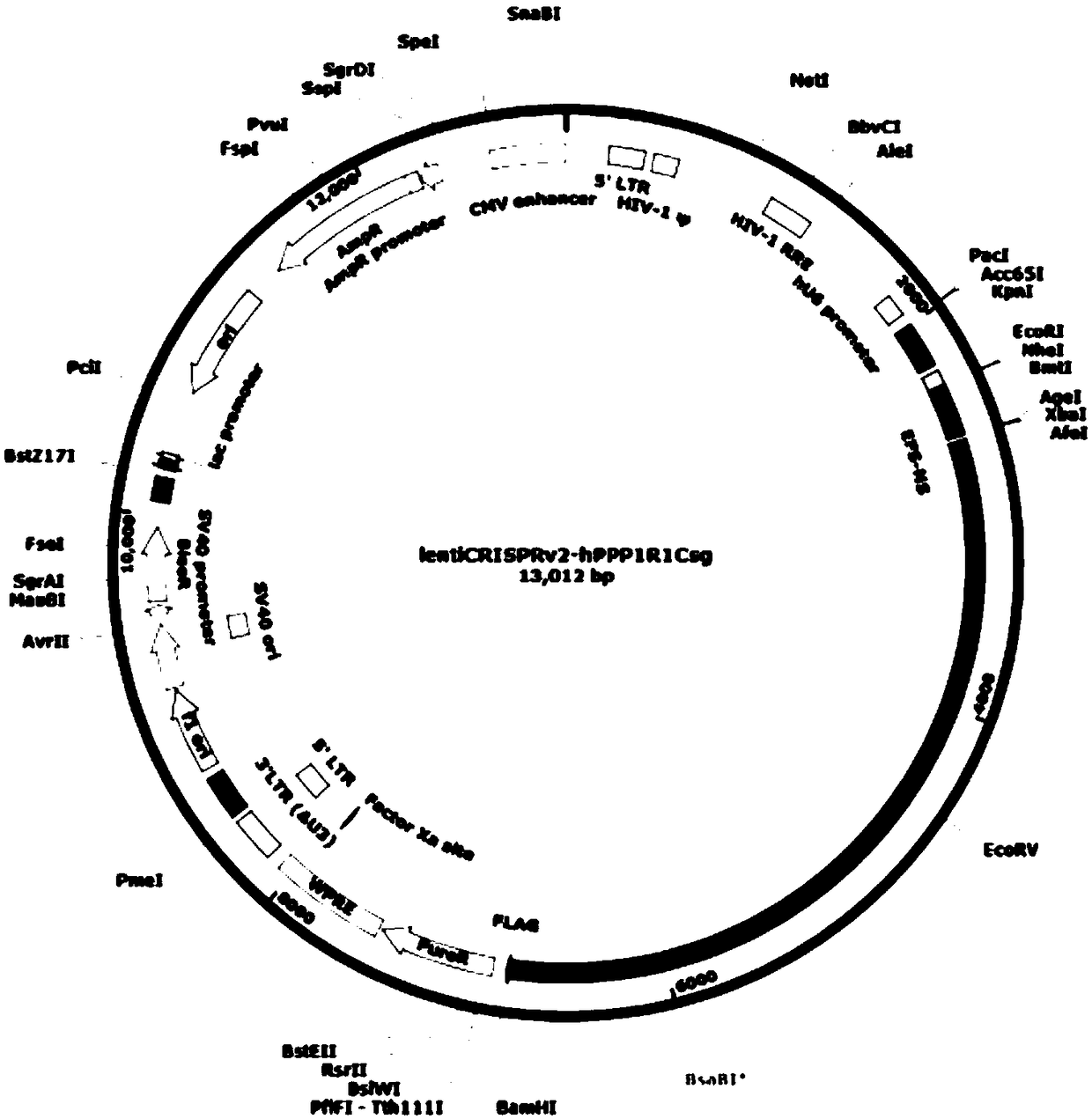
CRISPR-Cas9 targeted-knockout human colorectal cancer cell PPP1R1C gene and specific sgRNA thereof
The invention discloses a CRISPR-Cas9 targeted-knockout human colorectal cancer cell PPP1R1C gene and a specific sgRNA sequence. The invention comprises the following steps: firstly, acquiring sgRNA of a specific targeting PPP1R1C gene functional area, wherein a base sequence of the sgRNA is shown as SEQ ID NO.1; then, constructing an sgRNA-lentiviral vector system of the PPP1RQC gene, wherein thesystem contains Cas9 protein; and finally, infecting human colorectal cancer HT-29 cells with CRISPR / Cas9 lentivirus containing the sgRNA, so that a cell line that a PPP1R1C protein expression levelis obviously reduced is obtained. The invention is simple in operating steps, good in sgRNA targeting performance and high in cutting efficiency on the PPP1R1C gene; the constructed CRISPR / Cas9 lentiviral system is high in knockout efficiency and is capable of achieving specific knockout of the PPP1R1C gene, so that the human colorectal cancer cells that the PPP1R1C gene is knocked out is obtained; therefore, a powerful tool is provided for further researching an action mechanism of the PPP1R1C in the colorectal cancer cells.
Owner:OBIO TECH SHANGHAI CORP LTD
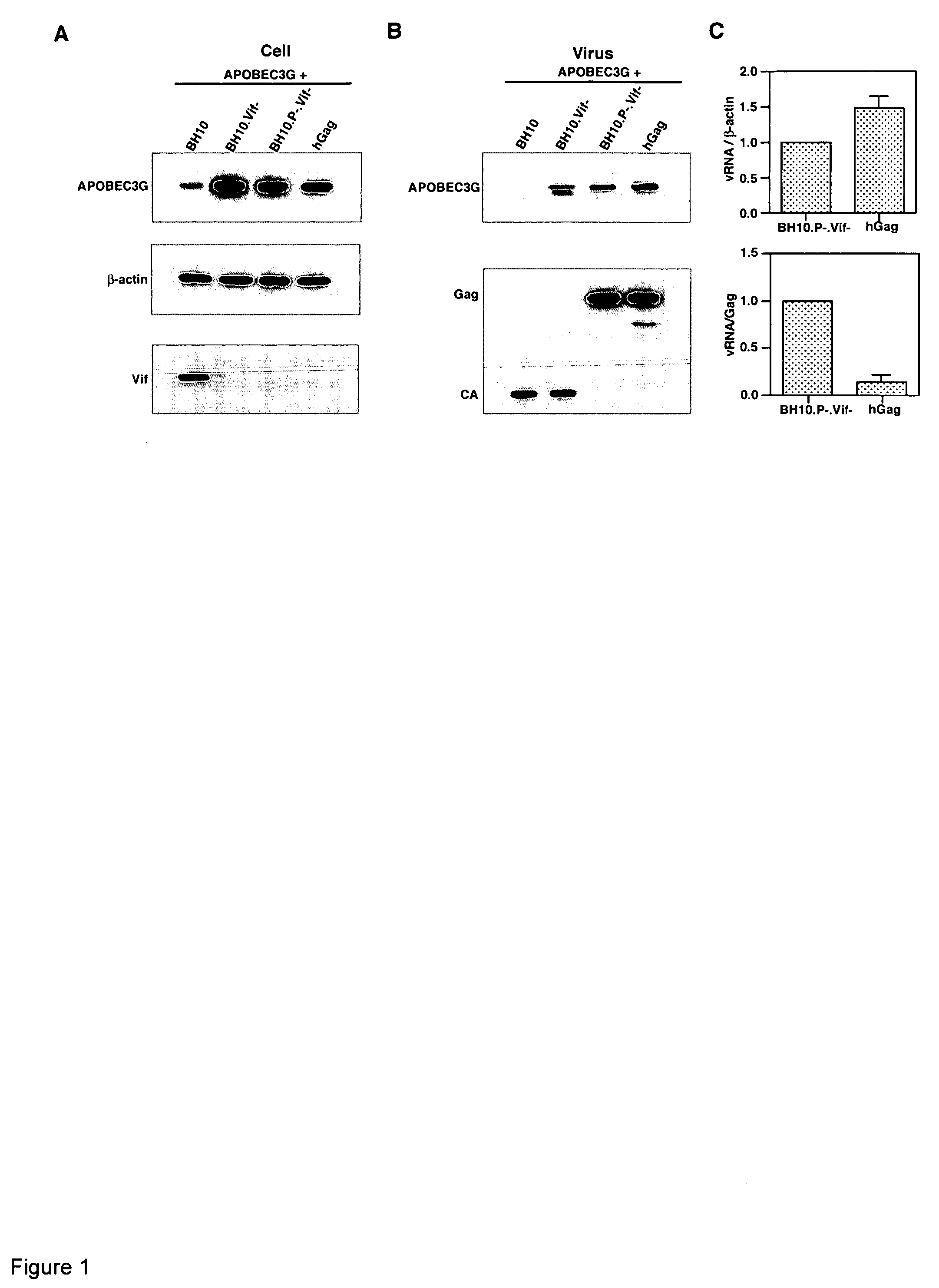
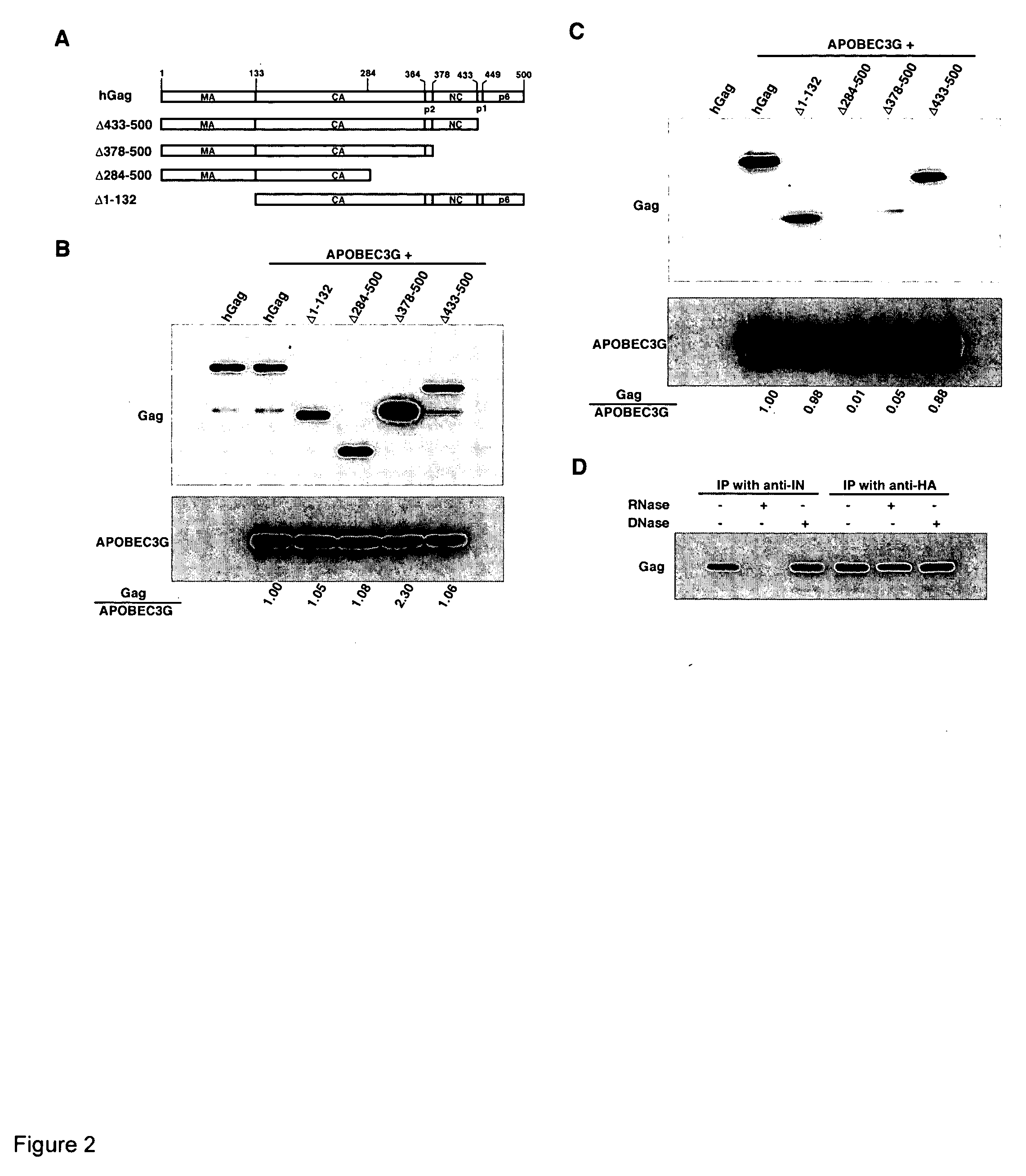
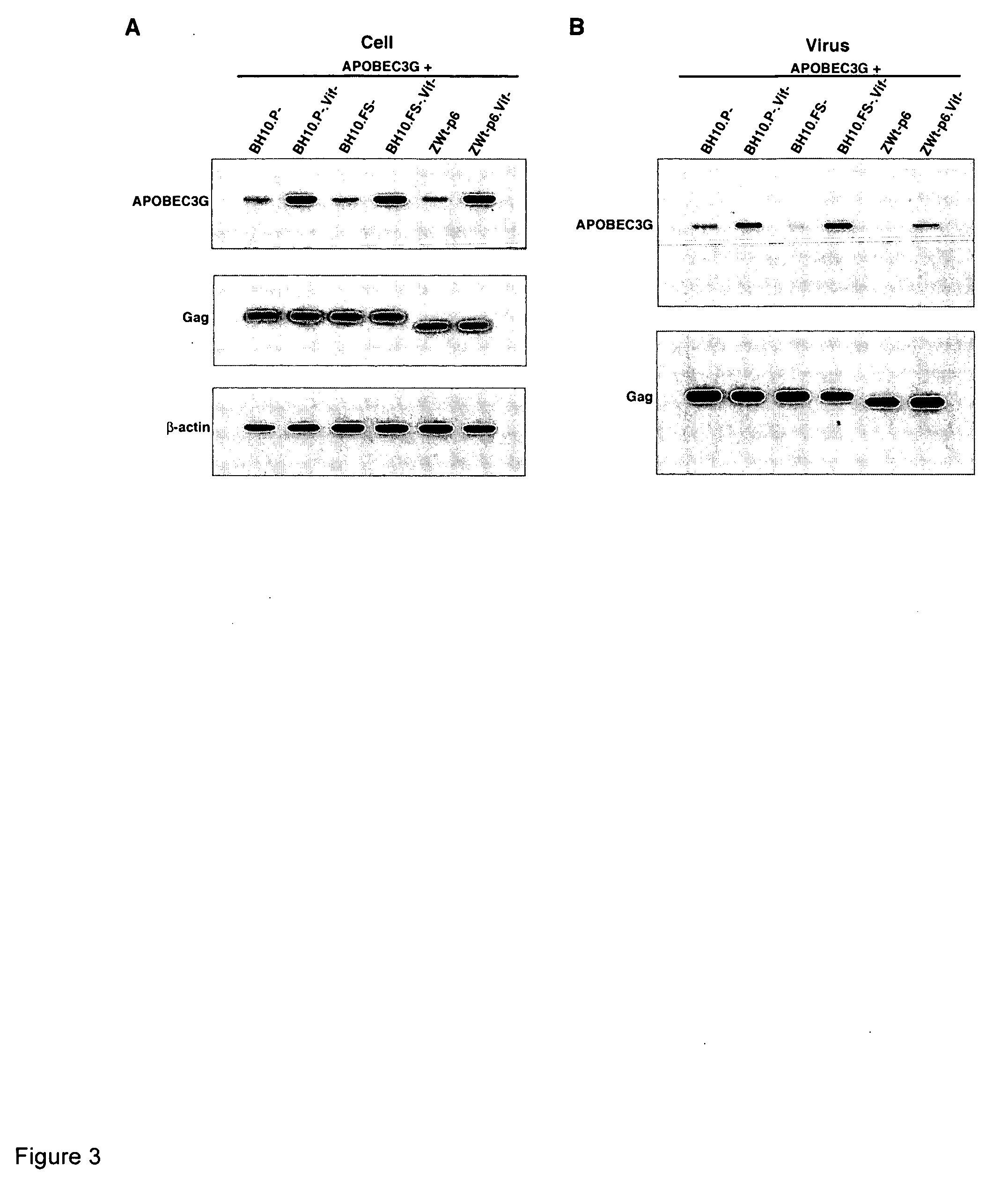
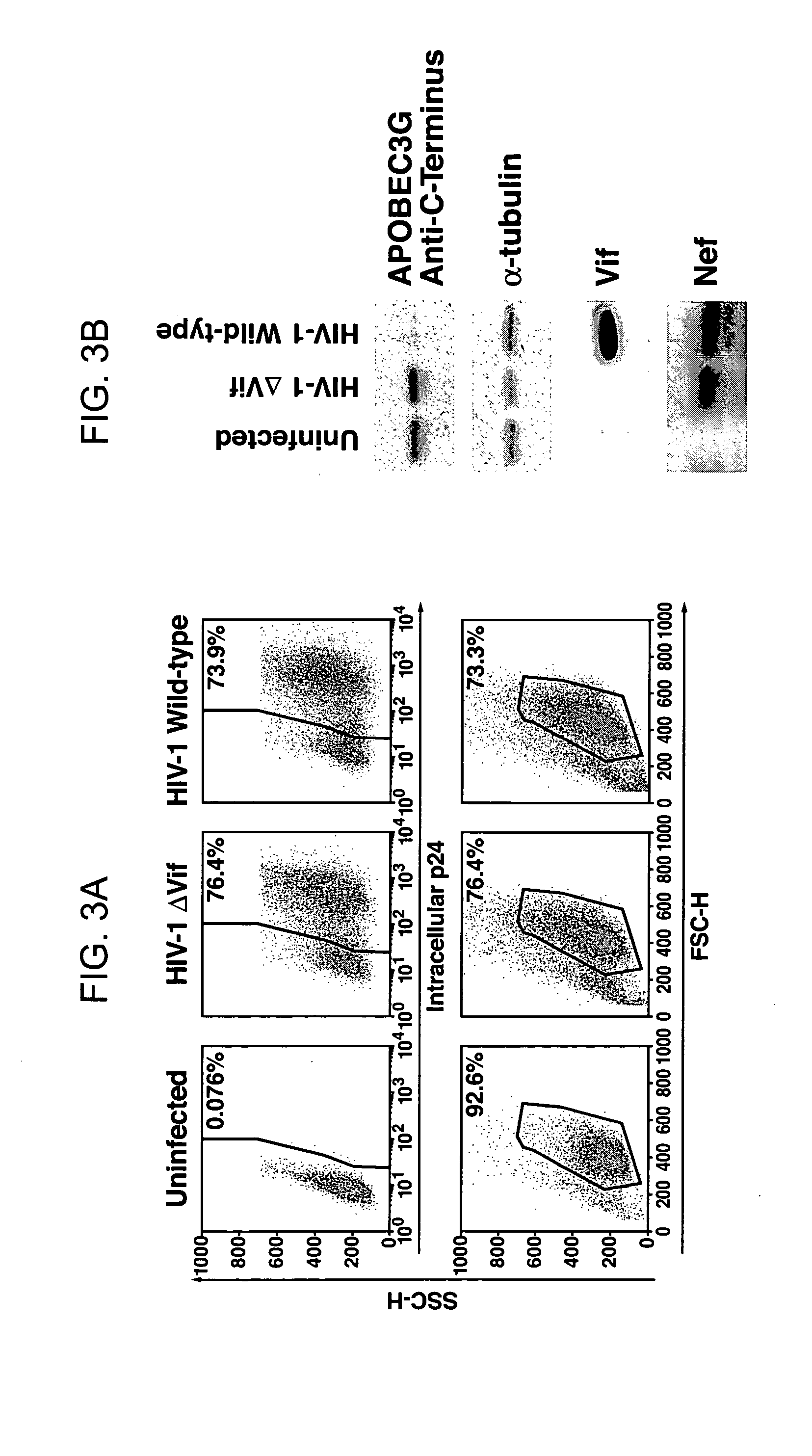
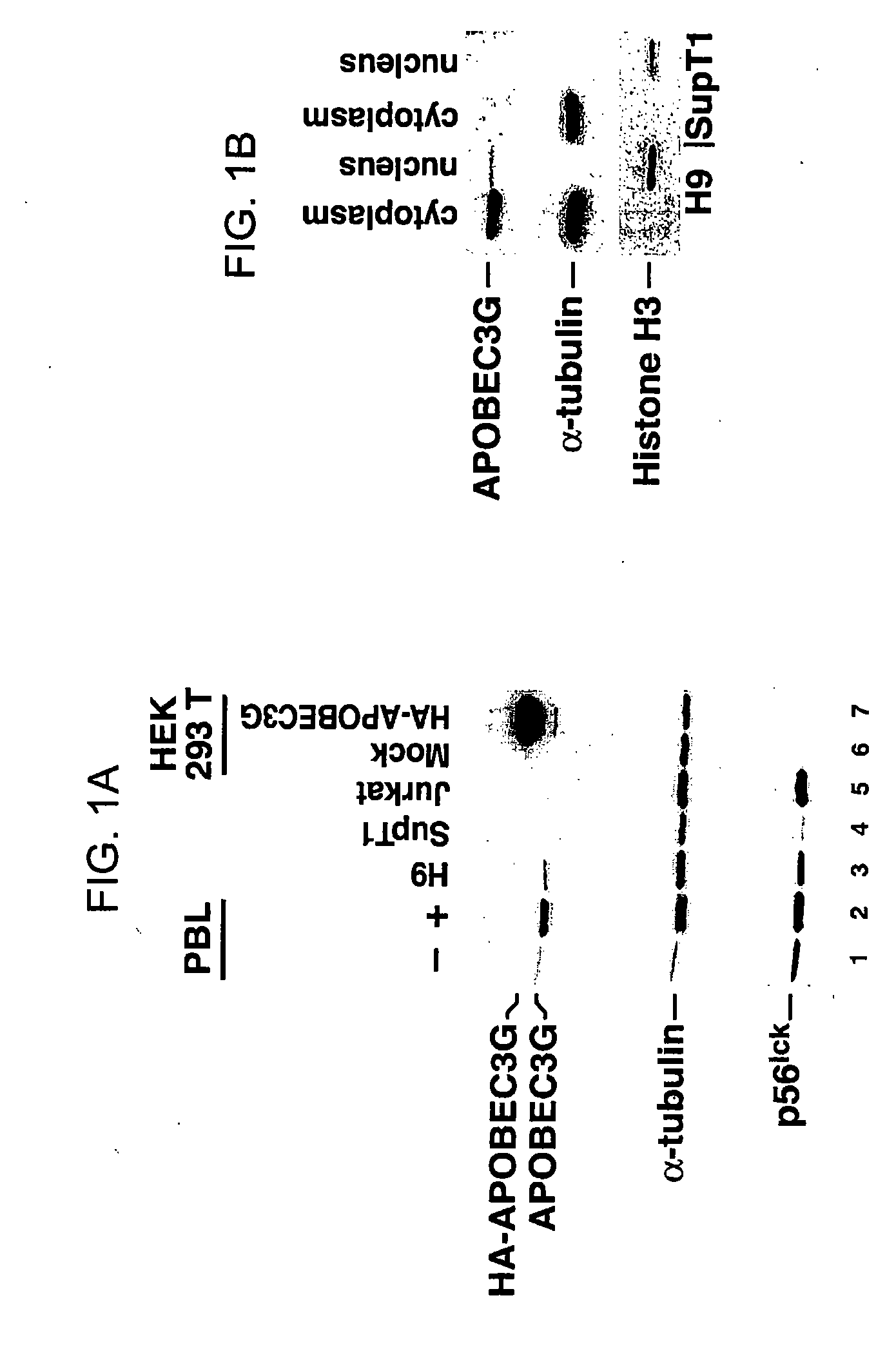
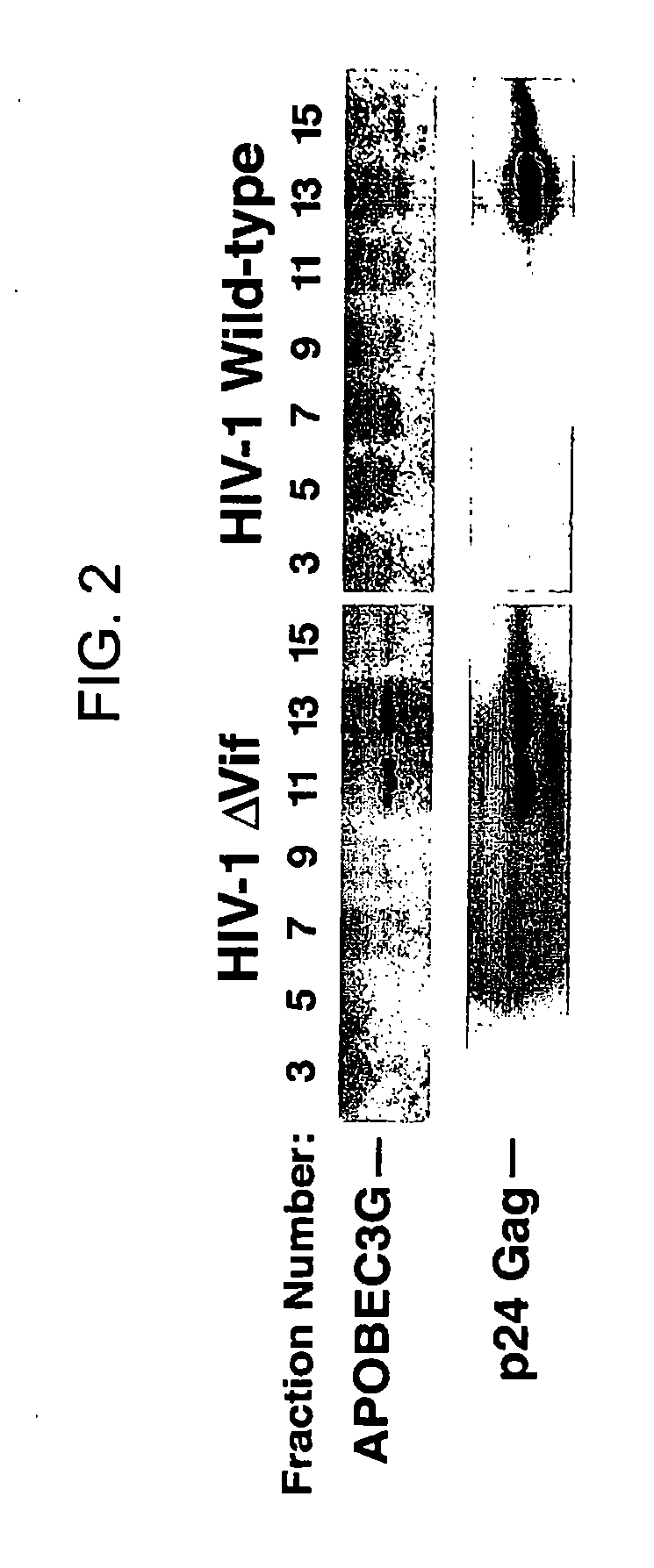
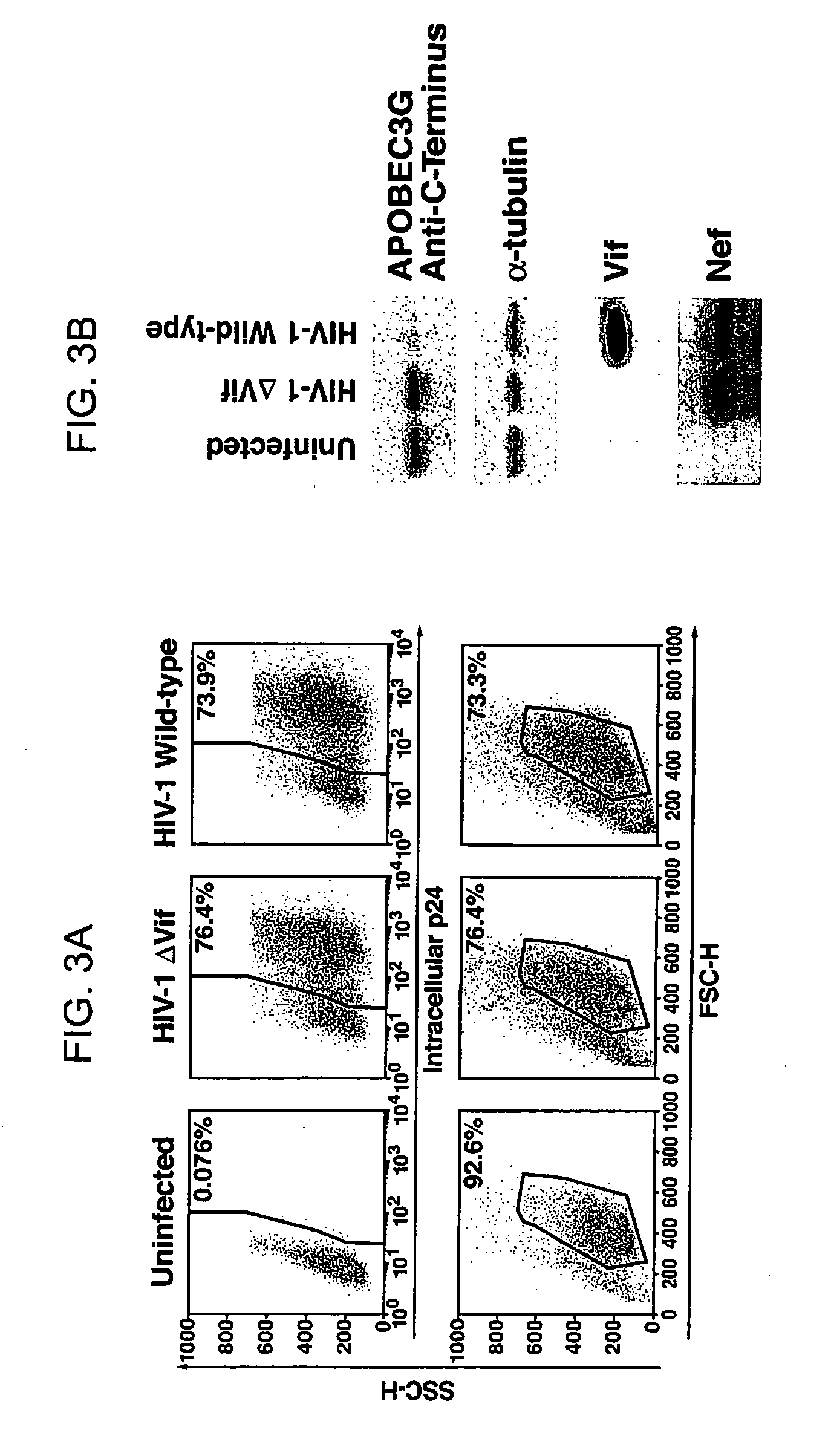
Inhibition of the tRNALys3-primed initiation of reverse transcription in HIV-1 by APOBEC3G
InactiveUS20060002951A1Inhibiting and reducing Vif-dependent inhibitionHigh affinityHydrolasesPeptide/protein ingredientsViral Reverse TranscriptionViral infection
The present invention generally relates to the field of antiviral therapy. More specifically, the present invention relates to the inhibition of the tRNALys3-primed initiation of reverse transcription in viruses by APOBEC3G. The present invention further relates to a method of treating or preventing viral infections by inhibiting tRNALys3 annealing and / or priming on a viral genome thereby reducing viral replication. More particularly, the present invention relates to the use of APOBEC3G, fragments or derivatives thereof for treatment or prophylaxis of HIV-1 infection and related lentivirus infections.
Owner:MCGILL UNIV
Composition and method for increasing cell density in cell cultures infected with lentivirus
InactiveUS7160721B2Increase cell densityViral antigen ingredientsMicrobiological testing/measurementBiotechnologyLentivirus
A composition and method for enhancing cell growth and increasing the density of cell cultures containing lentivirus-infected host cells comprises adding a suitable quantity of an antibiotic to the culture to destroy harmful bacteria.
Owner:ELANCO US INC
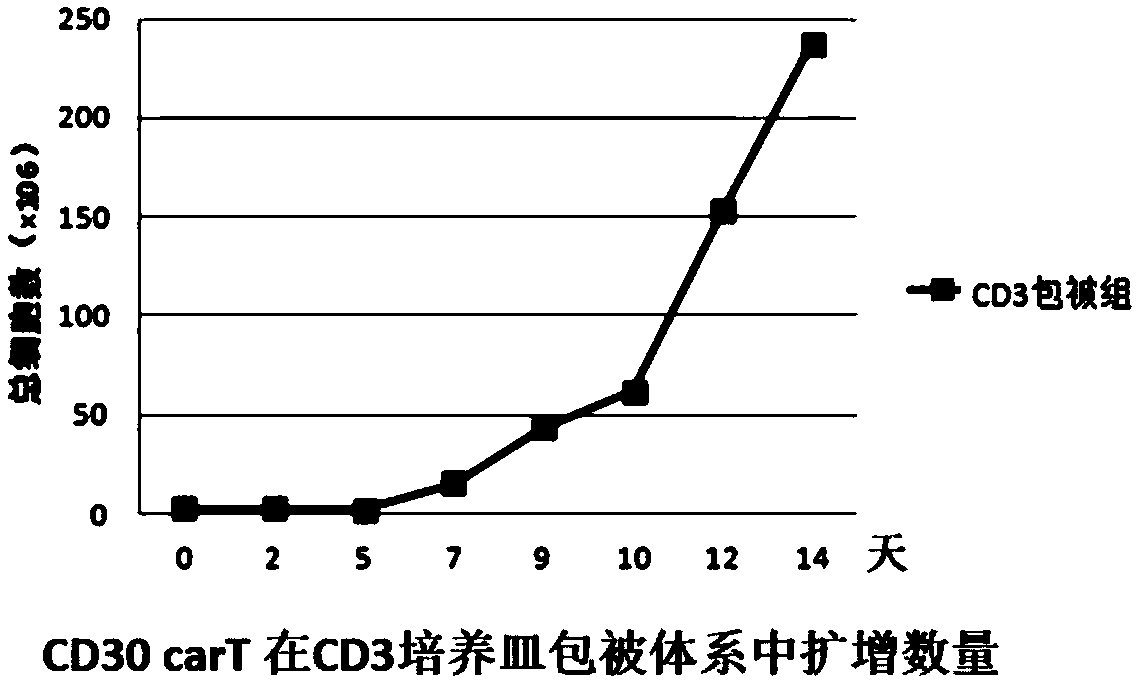
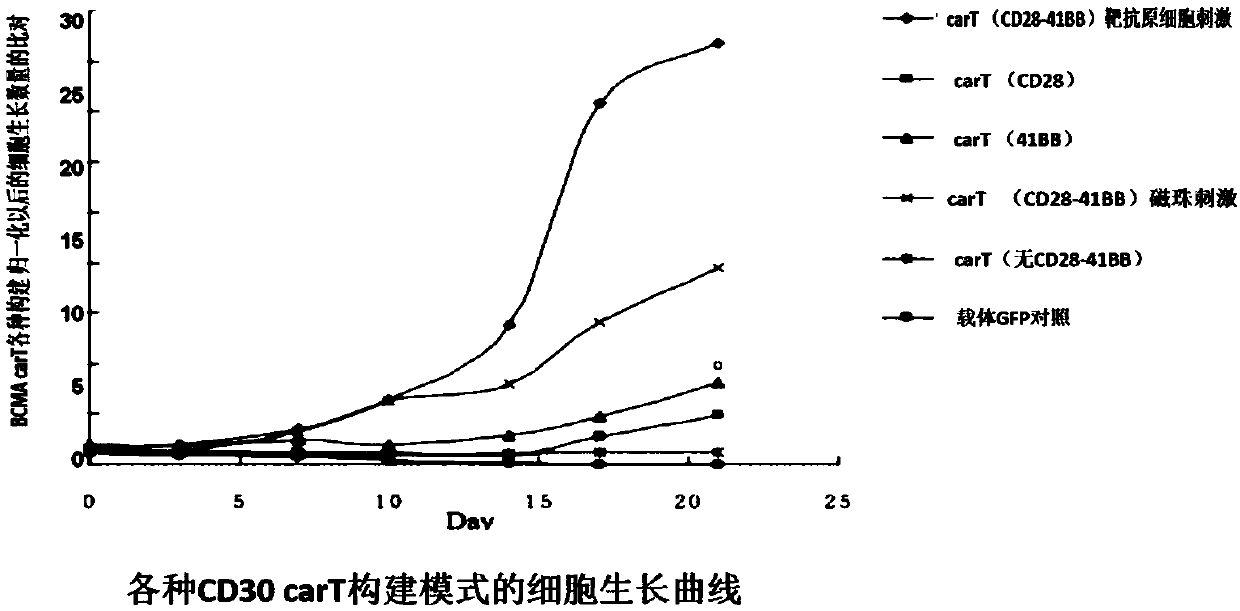
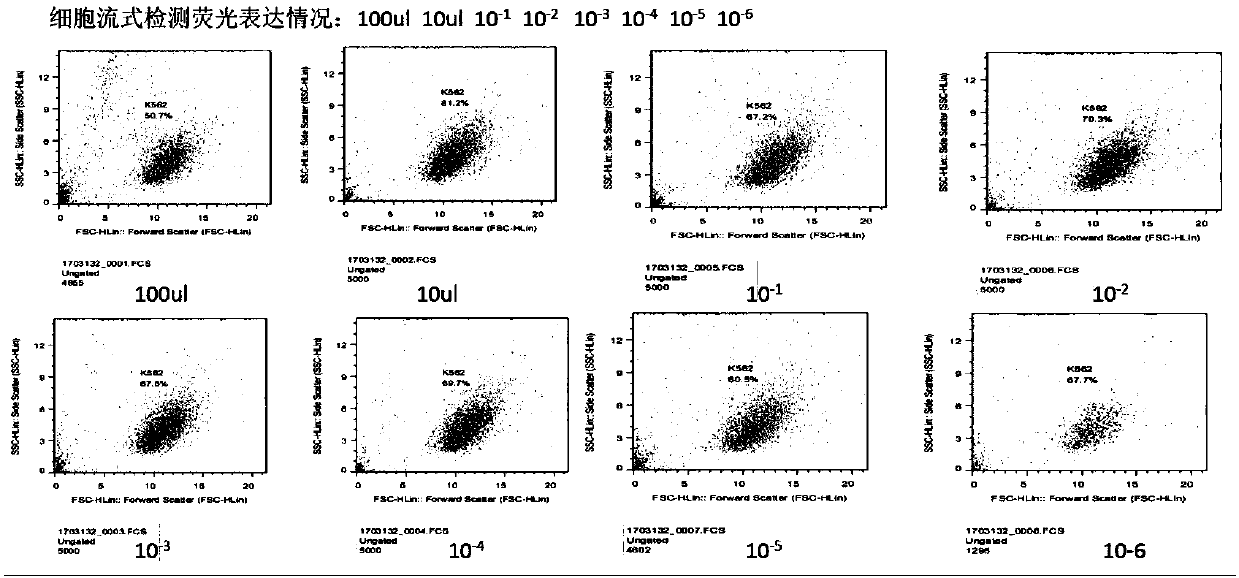
Transgenic T cell of targeted CD30 antigen as well as preparation method and application of transgenic T cell
InactiveCN107759699ADisinhibitionFunction increaseHydrolasesAntibody mimetics/scaffoldsAntigenPrimary cell
The invention discloses a transgenic T cell of a targeted CD30 antigen. The transgenic T cell is a primary cell which is integrated with a gene shown as SEQ ID NO:2 and encoding the targeted CD30 antigen, and knocks out a PD1 gene and / or CTLA4 gene, or is a primary cell containing a recombinant lentivirus expression vector (including a gene which is shown as SEQ ID NO:2 and encodes the targeted CD30 antigen and shRNA of a targeted PD1 gene or / and shRNA of a targeted CTLA4 gene); the primary cell is CD4+T cell or CD8+T cell. A preparation method comprises the following steps: firstly, carryingout lentivirus infection on the CD4+T cell or the CD8+T cell; secondly, mixing gRNA, CRISPR-cas9mRNA and HDR, and carrying out electroporation recombination on the T cell to obtain a finished product.According to the transgenic T cell disclosed by the invention, a recognition sequence of an EGFR (Epidermal Growth Factor Receptor) is introduced in carT construction; if necessary, a carT cell can be eliminated by using EGFR monoclonal antibody Cetuximab, the PD1 gene and the CTLA4 gene are knocked out or silenced, inhibition of the gene to the carT cell is eliminated, and the function of overcoming a tumor microenvironment and inhibiting immune cells by the carT cell are enhanced.
Owner:YINFENG BIOLOGICAL GRP
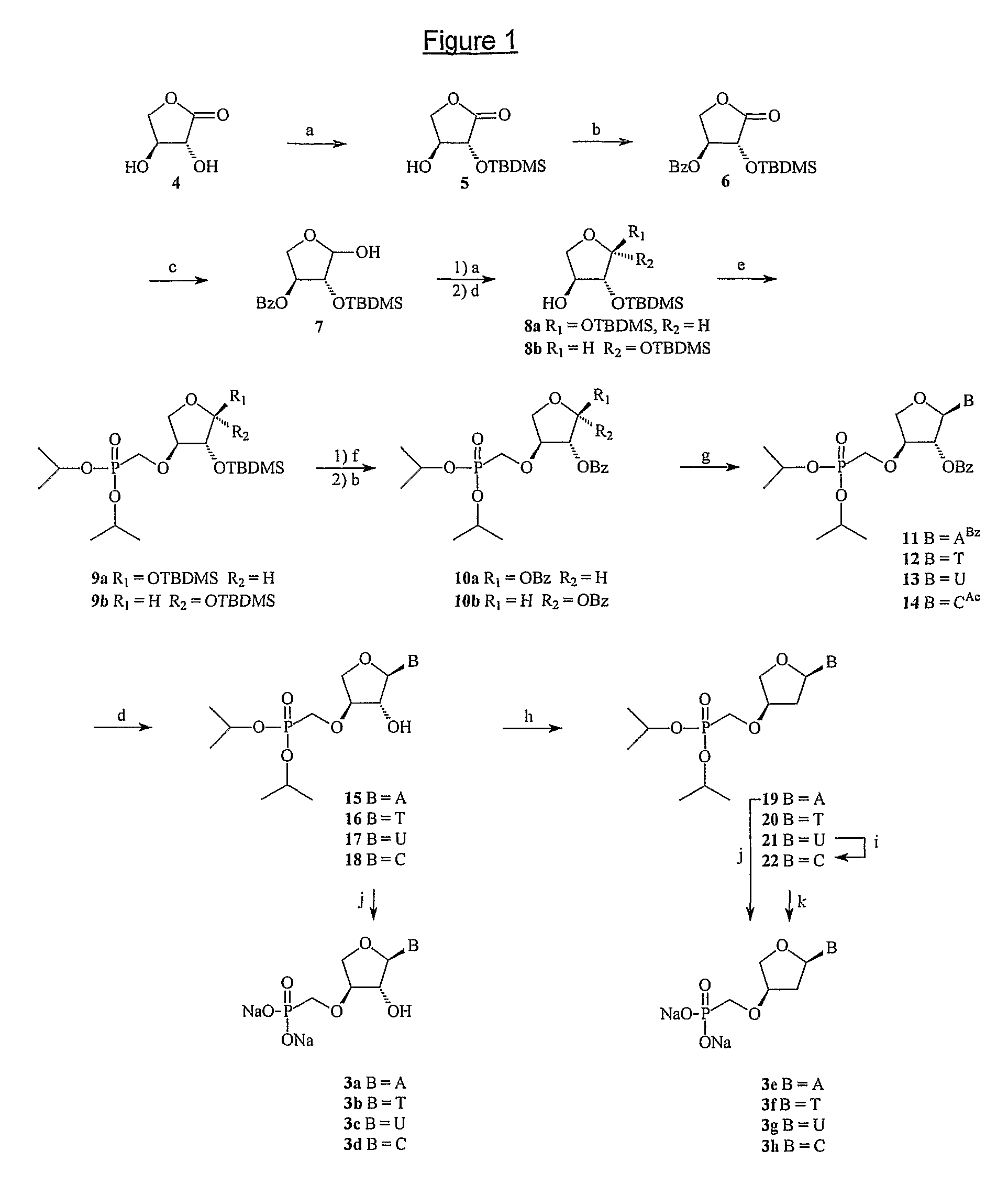
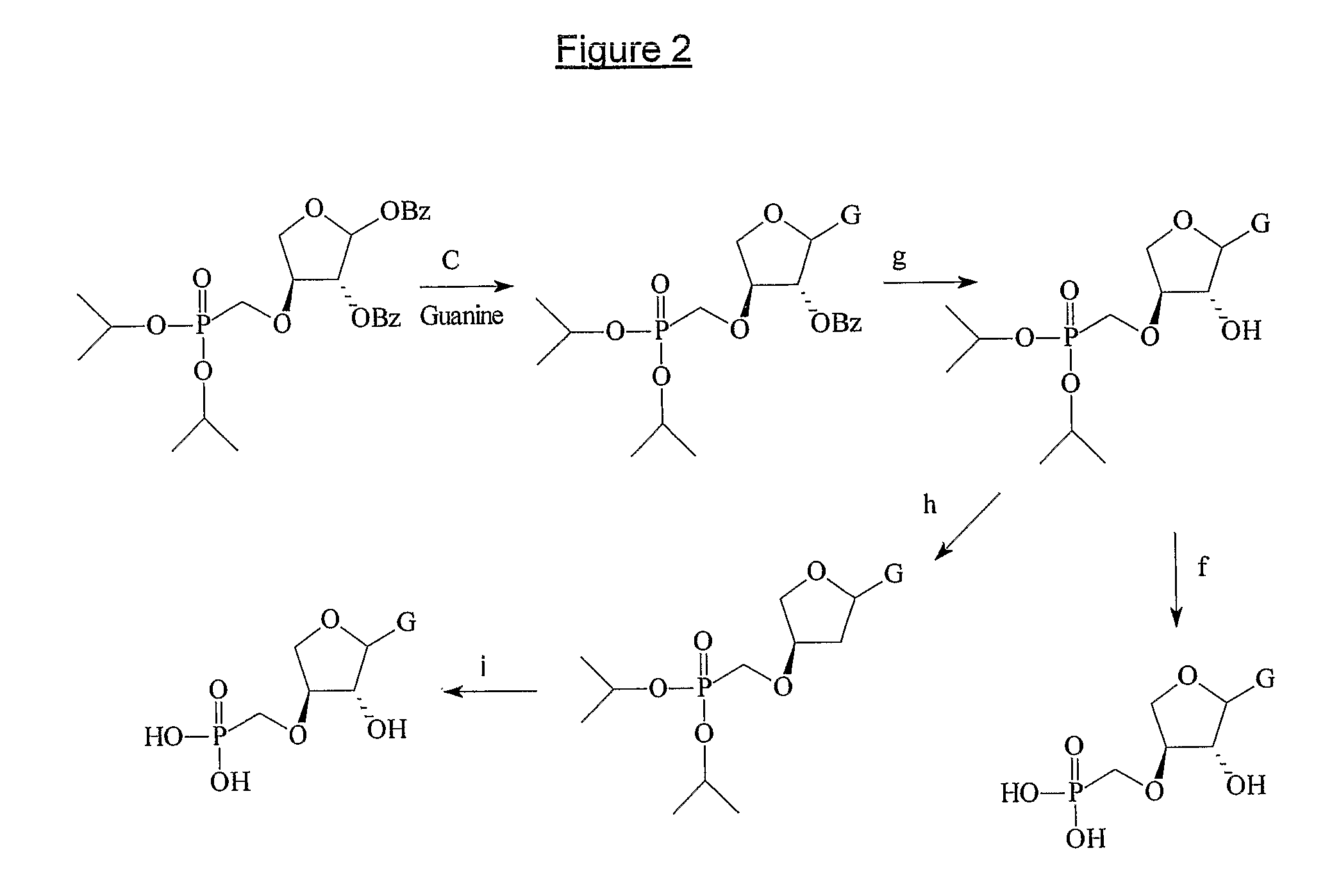
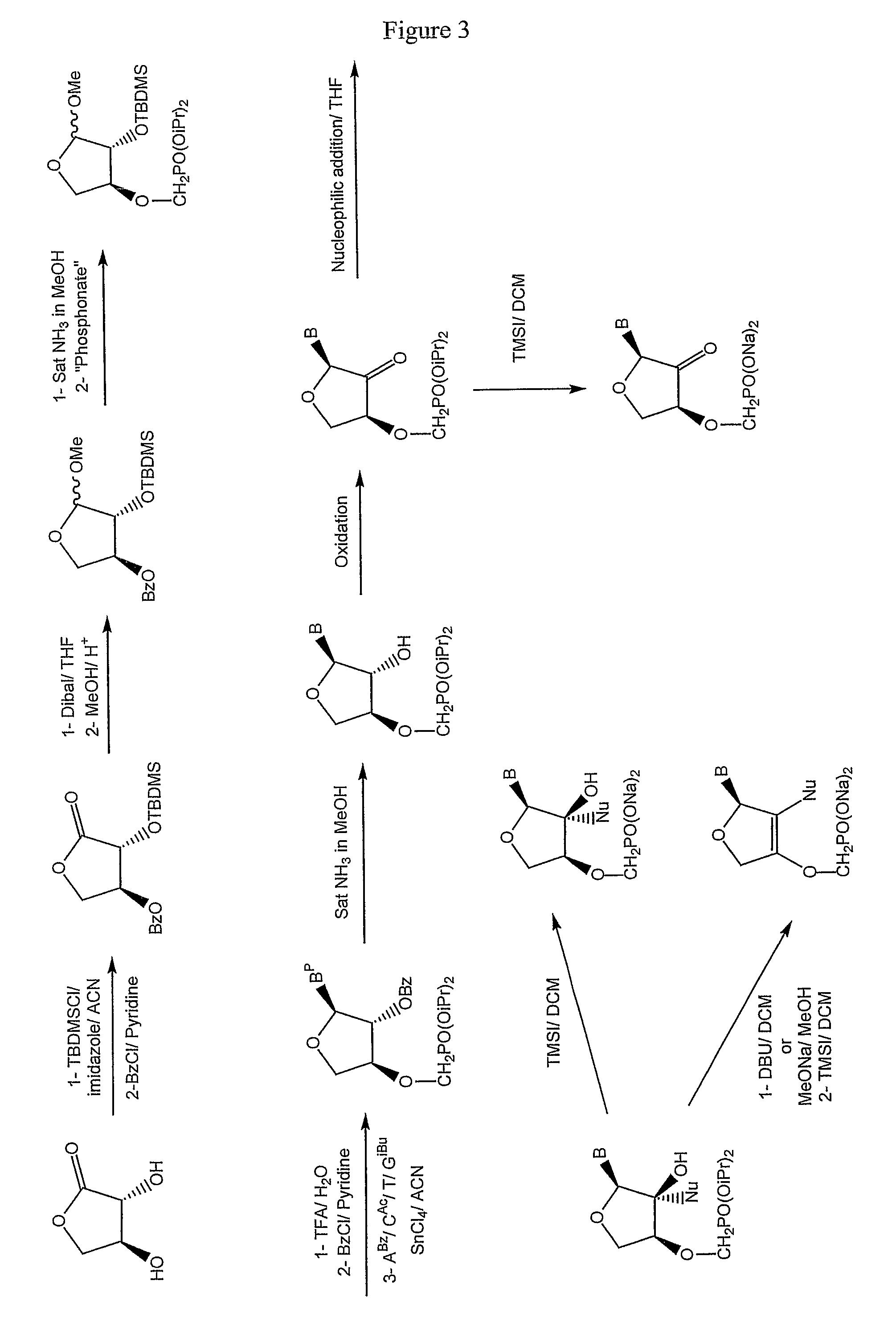
Phosphonate nucleosides useful as active ingredients in pharmaceutical compositions for the treatment of viral infections, and intermediates for their production
InactiveUS8076310B2Easy to tunePoor propertyBiocideAntibiotics chemistryNucleobaseBULK ACTIVE INGREDIENT
Disclosed herein are novel phosphonate nucleosides and thiophosphonate nucleosides comprising a phosphonalkoxy-substituted or phosphonothioalkyl-substituted five-membered, saturated or unsaturated, oxygen-containing or sulfur-containing ring coupled to a heterocyclic nucleobase such as a pyrimidine or purine base. The invention further relates to compounds having HIV (Human Immunodeficiency Virus) replication inhibiting properties and to compounds having antiviral activities with respect to other viruses. The invention also relates to methods for preparation of all such compounds and pharmaceutical compositions comprising them. The invention further relates to the use of said compounds as a medicine and in the manufacture of a medicament useful for the treatment of subjects suffering from HIV infection, as well as for treatment of other viral, retroviral or lentiviral infections and to the treatment of animals suffering from FIV, viral, retroviral or lentiviral infections.
Owner:K U LEUVEN RES & DEV
CAR-NK cell as well as preparation method and application thereof
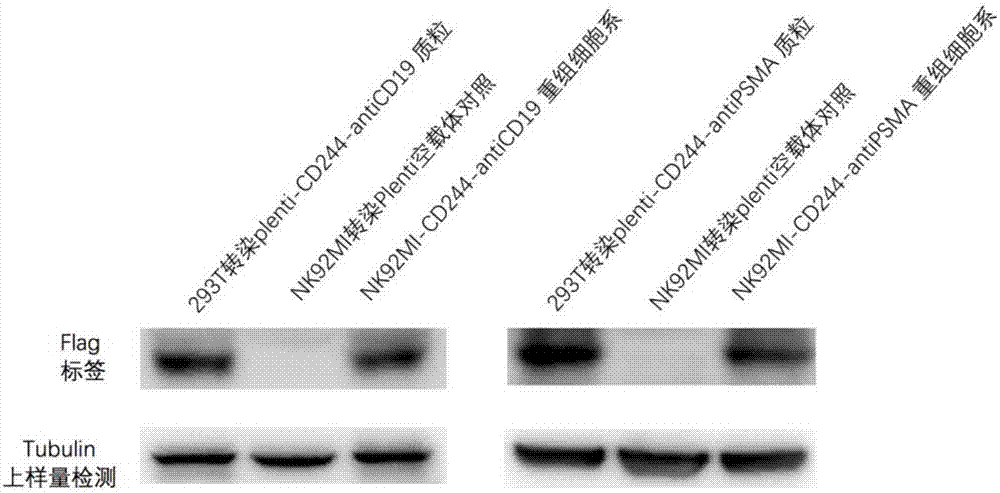
ActiveCN107034237AInduced lysisReduce manufacturing costMammal material medical ingredientsNucleic acid vectorCAR PROTEINLentivirus
The invention relates to a CAR-NK cell as well as a preparation method and an application thereof. The method comprises the following steps: S1, linking a specific antibody coding gene into a cloning site region of a modified CAR skeleton so as to obtain a CAR gene, wherein the modified CAR skeleton comprises a CD244 episporium signal region, the cloning site region, a CD244 extracellular hinge region, a CD244 transmembrane part region and a CD244 intracellular part region which are sequentially connected; and S2, infecting the CAR gene with NK cell through lentivirus, so that the NK cell expressing the CAR protein is obtained, namely the CAR-NK cell. The prepared tumor immnuotherapy cell, namely the CAR-NK cell, of the invention, besides a patient with cancer, is also applicable to allosome infusion; therefore, the prepared tumor immnuotherapy cell, in comparison with existing CAR-T, is wider in application scope and is lower in preparation cost.
Owner:国健亦诺生物科技(北京)有限公司
Preparation method and application of CIK (Cytokine-induced Killer) modified by anti-human CD19 chimeric antigen receptor
ActiveCN106544365AStrong killing effectProliferation effect is goodGenetically modified cellsMammal material medical ingredientsAntigen receptorLentivirus
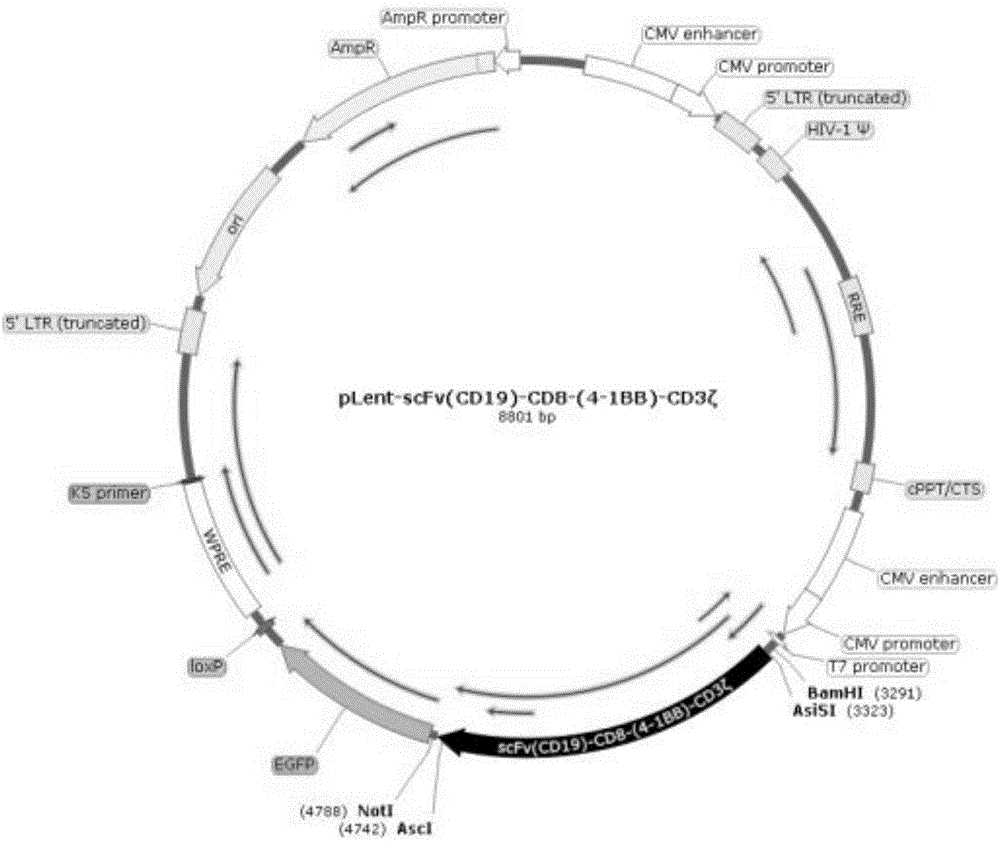
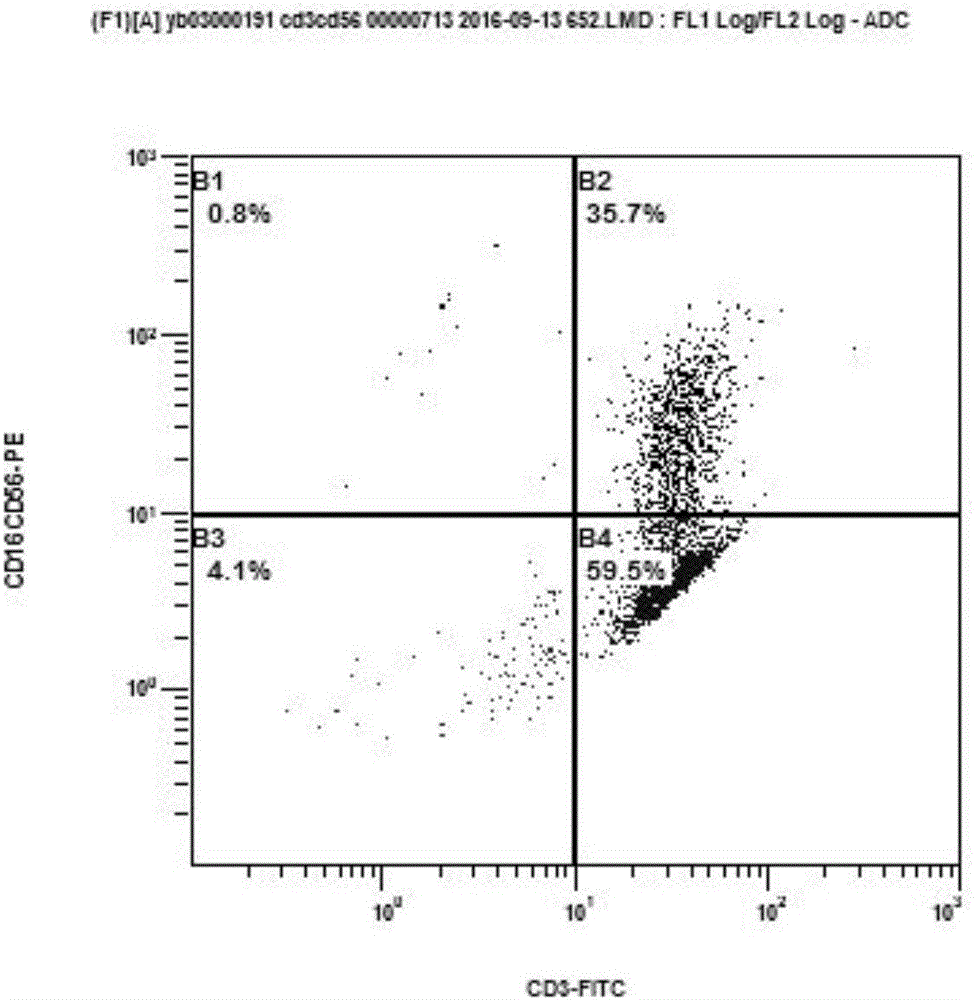
The invention discloses a preparation method and application of a CIK (Cytokine-induced Killer) modified by an anti-human CD19 chimeric antigen receptor. The preparation method comprises the steps of coding a fusion gene segment of the chimeric antigen receptor scFv (CD19)-CD8-(4-1BB)-CD3 zeta; inserting the fusion gene segment into a lentiviral expression vector; packaging into a lentivirus carried with a scFv (CD19)-CD8-(4-1BB)-CD3 zeta coding gene; and infecting the CIK induced by autologous lymphocytes of a patient on the lentivirus carried with the scFv (CD19)-CD8-(4-1BB)-CD3 zeta coding gene. The CIK modified by the chimeric antigen receptor scFv (CD19)-CD8-(4-1BB)-CD3 zeta can be applied in treatment of B cell lymphoma leukemia.
Owner:SHANDONG XINRUI BIOTECH CO LTD
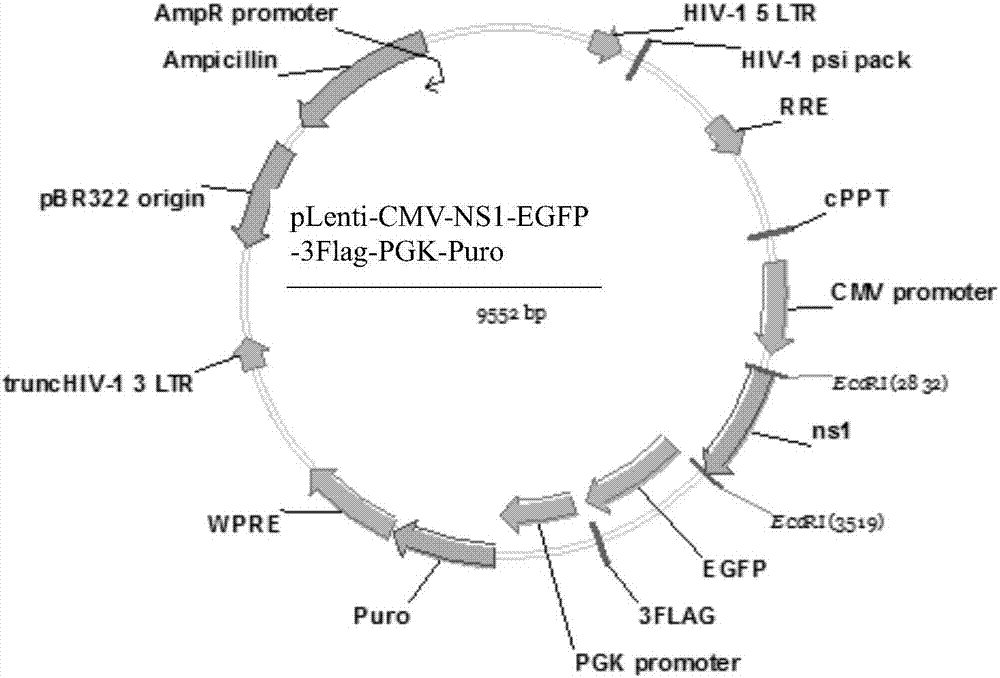
Building method of cell strain for stably expressing NS1(non-structural 1) protein
InactiveCN107190024AEasy to detectSsRNA viruses negative-senseVirus peptidesImmunofluorescenceWestern blot
The invention relates to a building method of a cell strain for stably expressing NS1 (non-structural 1) protein. An NS1 gene is inserted in an EcoR I polyclone site of a pLenti-CMV-EGFP-3Flag-PGK-Puro carrier; a built pLenti-CMV-NS1-EGFP-3Flag-PGK-Puro recombinant plasmid and a virus packaging plasmid are used for 293T cell cotransfection; culture, toxin elimination, virus liquid supernatant collection and filtering are performed to obtain recombinant lentivirus liquid. Packaged recombinant lentivirus infection A549 cells are subjected to puromycin screening, immunofluorescence and Western blot identification to obtain the cell strain for stably expressing NS1 protein. A recombinant lentivirus system is used for preparing the cell strain fusing tag protein capable of stably expressing the NS1 protein. The cell strain provides a good tool for studying the biological function of the NS1 protein.
Owner:LIAONING UNIVERSITY
Methods for treating lentivirus infections
InactiveUS20050053977A1Improve the level ofInhibitory activityCompound screeningApoptosis detectionScreening methodVif Protein
The present invention provides methods of identifying an agent that inhibits an activity of a lentiviral Vif protein. The present invention provides methods of identifying an agent that increases the level of active APOBEC3G in a cell. The present invention provides agents identified by a subject screening method; and further provides methods for treating lentivirus infections.
Owner:THE J DAVID GLADSTONE INST A TESTAMENTARY TRUST ESTABLISHED UNDER THE WILL OF J DAVID GLADS
Preparation method for efficiently amplifying NK cells by utilizing trophoblasts
ActiveCN110684730APromote activationImprove build success rateGenetically modified cellsBlood/immune system cellsNatural Killer Cell Inhibitory ReceptorsPlasmid Vector
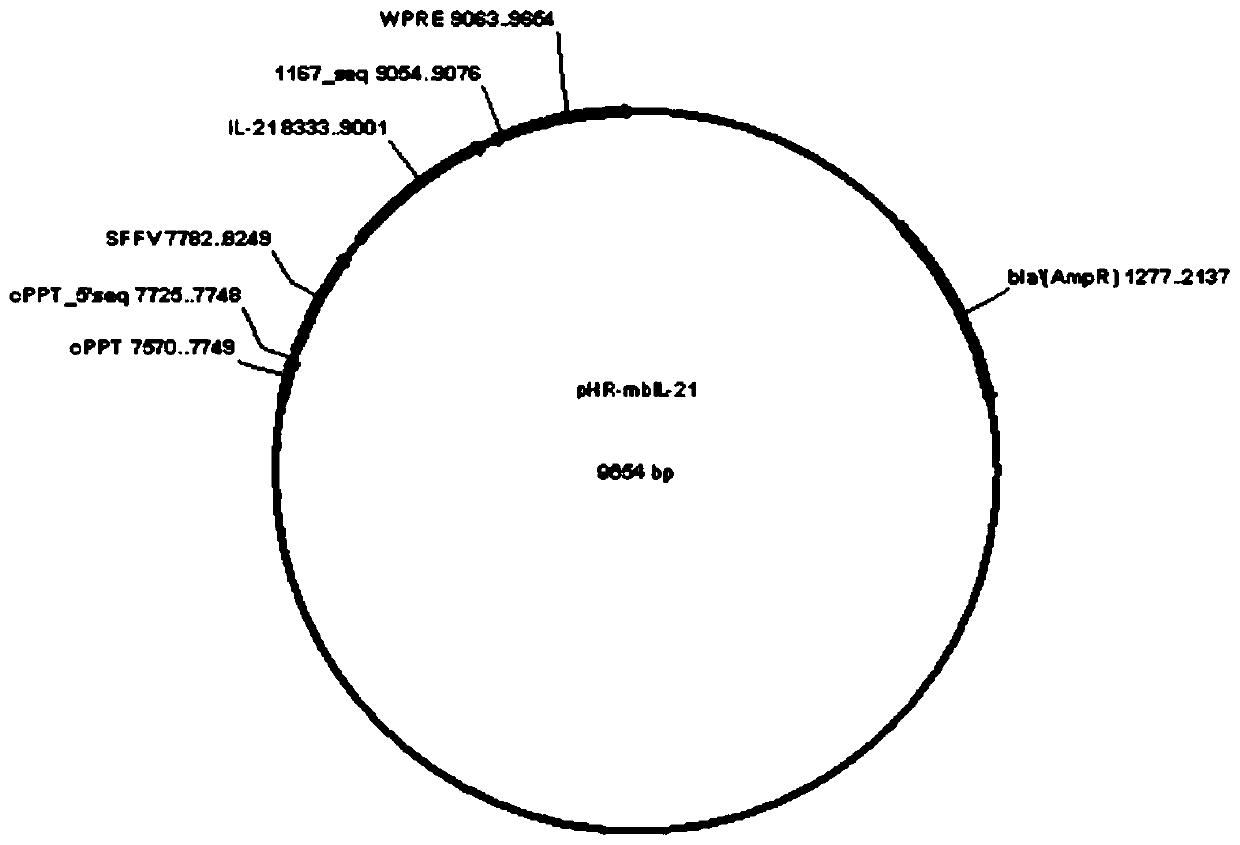
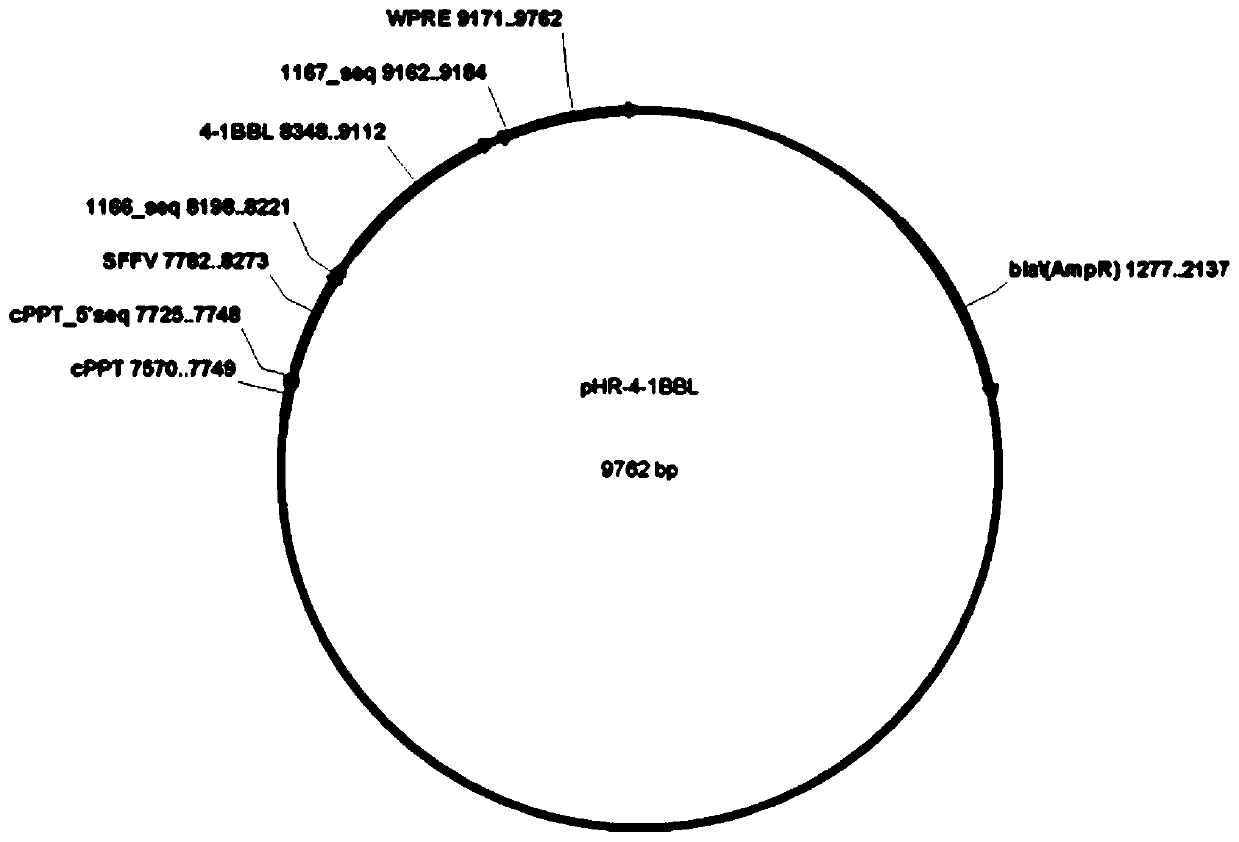
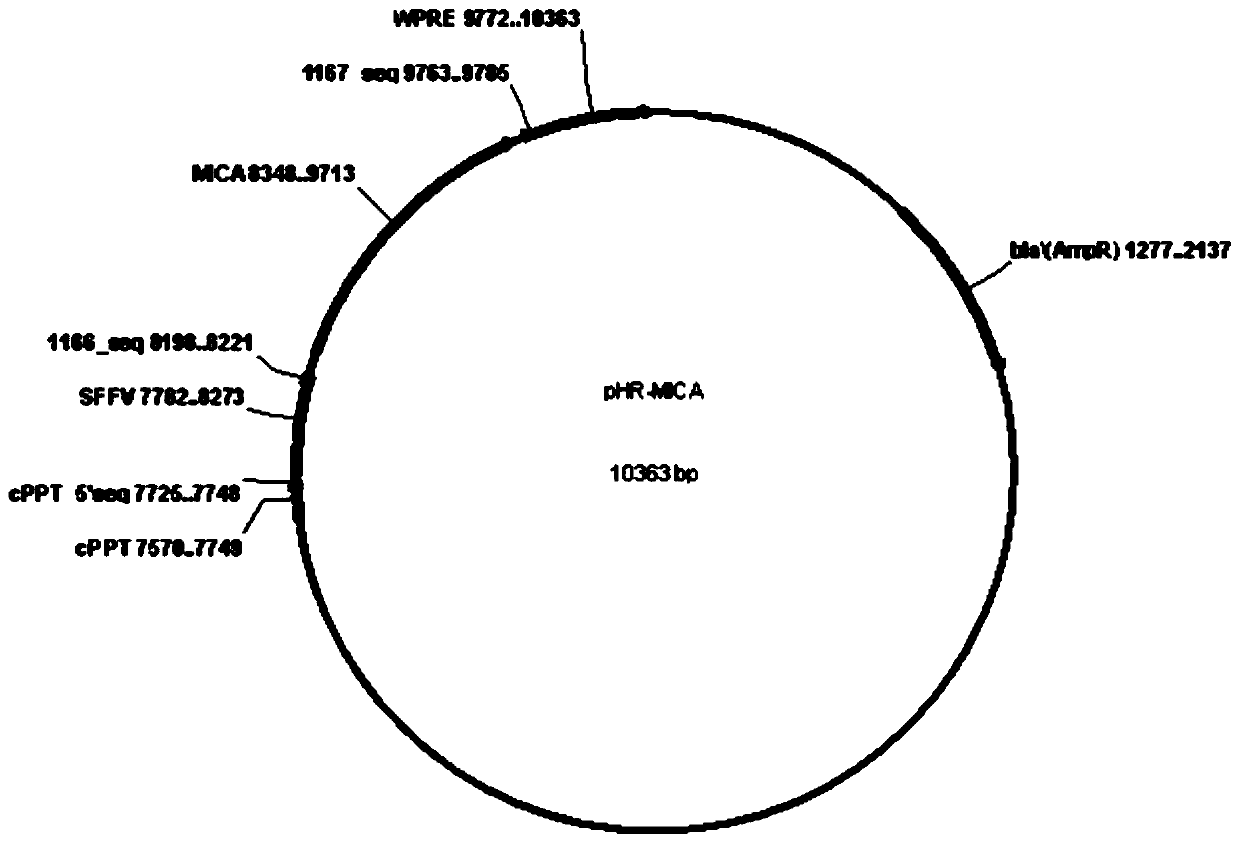
The invention relates to the field of gene engineering and cytobiology, in particular to a preparation method for efficiently amplifying NK cells by utilizing trophoblasts. The construction method comprises the following steps: (1) constructing a pHR-mbIL-21 plasmid vector, a pHR-4-1BBL plasmid vector and a pHR-MICA plasmid vector; (2) preparing recombinant lentiviruses by using pHR-mbIL-21, pHR-4-1BBL and pHR-MICA plasmid vectors respectively; (3) preparing K562-mbIL-21-4-1BBL-MICA trophoblasts; (4) extracting PBMC cells; and (5) carrying out in-vitro amplification on NK cells. The inventionprovides the preparation method of NK cells. The method employs the step of independently constructing plasmid vectors for expressing mbIL-21, 4-1BBL and MICA molecules, K562 is infected with recombinant lentivirus, the NK cells can be prepared, the preparation method overcomes the defects in the prior art, the K562 cells simultaneously expressed by IL-21, 4-1BBL and MICA molecules are used as trophoblasts for amplification culture of the NK cells, the amplification multiple of the NK cells reaches up to 890 times, the purity of the prepared NK cells reaches 92.2%, and the repeatability of theamplification multiple between different PBMC cells is good.
Owner:山东德升生物工程有限公司
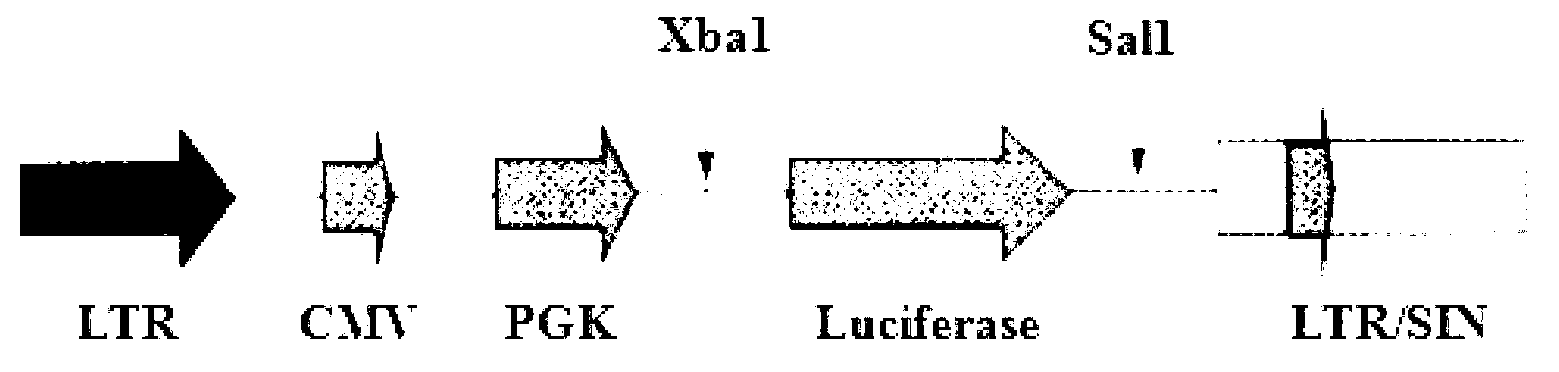
A549 nude mouse model of stably expressed luciferase and building and application thereof
InactiveCN102936600AObservation is intuitive and convenientReliable resultsGenetic engineeringFermentationWilms' tumorLentivirus Infections
The invention relates to an A549 nude mouse model of stably expressed luciferase and building and application thereof. The nude mouse model leads firefly luciferase genes to human lung adenocarcinoma A549 cell strains through the gene recombination technology and lentivirus infection, tumor clone cell strains of the stably expressed luciferase are obtained through subcloning screening, and recombinational A549 cells are inoculated in a mouse to obtain the A549 nude mouse model. Compared with a traditional tumor model drug effect detection method, the A549 nude mouse model enables observation to be visual and convenient, can be used for somatoscopy without damaging animals, is reliable in result, and has good application prospect.
Owner:SUZHOU RES INST OF TONGJI UNIV
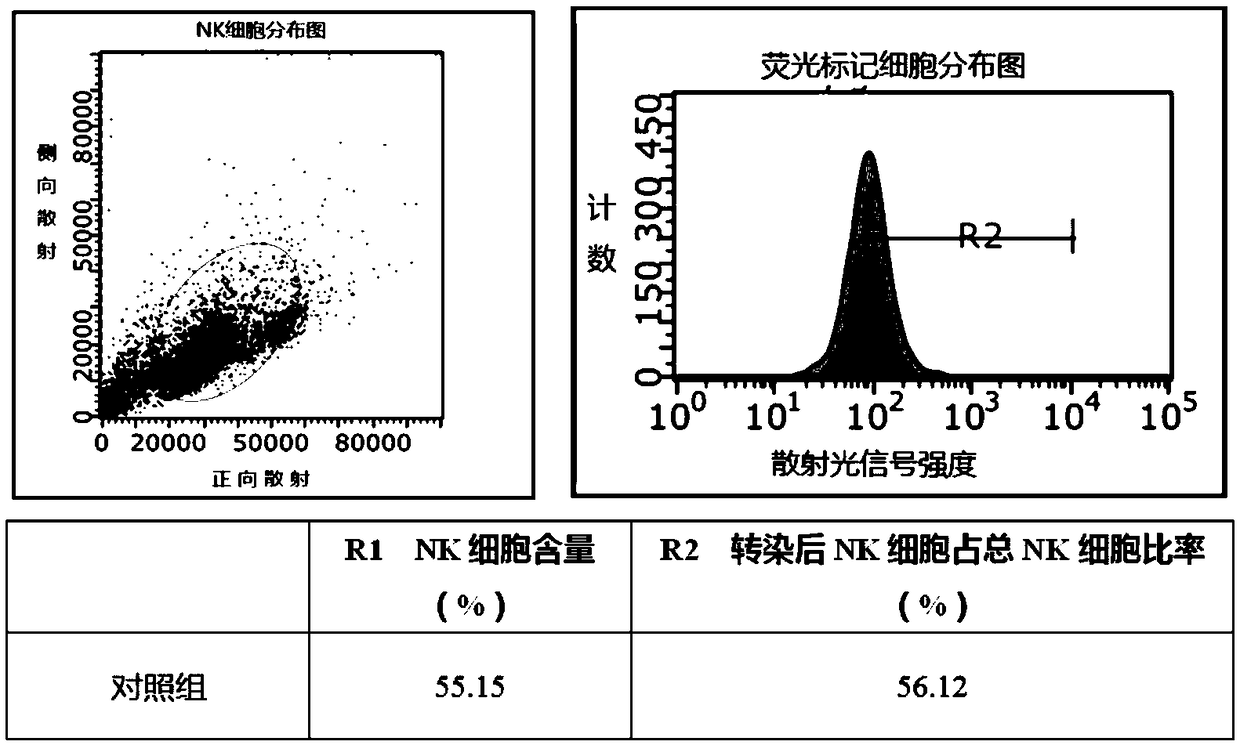
NK cell transfection efficiency improving method
ActiveCN108893493AHigh transfection efficiencyHigh infection efficiencyFermentationVector-based foreign material introductionNatural Killer Cell Inhibitory ReceptorsMicrobiology
The invention relates to the technical field of NK cell in-vitro amplification, in particular to a NK cell transfection efficiency improving method. In lentivirus infection of NK cells, an auxiliary infection agent 1 is added while a CAR-containing lentivirus carrier is added in primary infection; an auxiliary infection agent 2, an auxiliary infection agent 3 and an auxiliary infection agent 4 areadded while a CAR-containing lentivirus carrier is added in secondary infection; the auxiliary infection agent 1 refers to polybrene solution, the auxiliary infection agent 2 refers to IL2 solution,the auxiliary infection agent 3 refers to IL12 solution, and the auxiliary infection agent 4 refers to PHA solution. According to a Car-NK transfection method, different auxiliary infection agents areadded in a transfection process, NK cell transfection efficiency is imiproved, and NK cell lentivirus transfection efficiency can be improved from less than 10% to 50%-60%.
Owner:国健呈诺生物科技(北京)有限公司
Immortalized yak rumen epithelial cell line and construction method thereof
ActiveCN114058592AMethods for Enriching Isolation and CultureProliferation in good shapeGastrointestinal cellsVirus peptidesNutritionSignalling pathways
The invention discloses an immortalized yak rumen epithelial cell line and a construction method thereof. The preservation number of the cell line is CCTCC No. C2021245, and the cell line is classified and named as the immortalized yak rumen epithelial cell line SV40T-YREC-hTERT. The method comprises the following steps: performing adherent culture on tissue digested by primary yak rumen epithelial papilla type I collagenase to obtain primary rumen epithelial cells; with the cells as host cells, infecting the cells with lentiviruses carrying SV40T and hTERT genes; and performing screening and enlarged culture to obtain the immortalized yak rumen epithelial cell line. Through the establishment of the cell line, the defects that the primary rumen epithelial cells are difficult to culture and the number of passage times is limited are overcome, and yak source cell line resources are enriched; and the cell line can be stably passaged, can keep the morphological characteristics and normal functions of the primary rumen epithelial cells, and provides a useful in-vitro cell model for researching rumen epithelial nutrition digestion, absorption and metabolism, cell signal path mechanisms and the like.
Owner:SICHUAN AGRI UNIV
Immunomodulating compositions and uses therefor
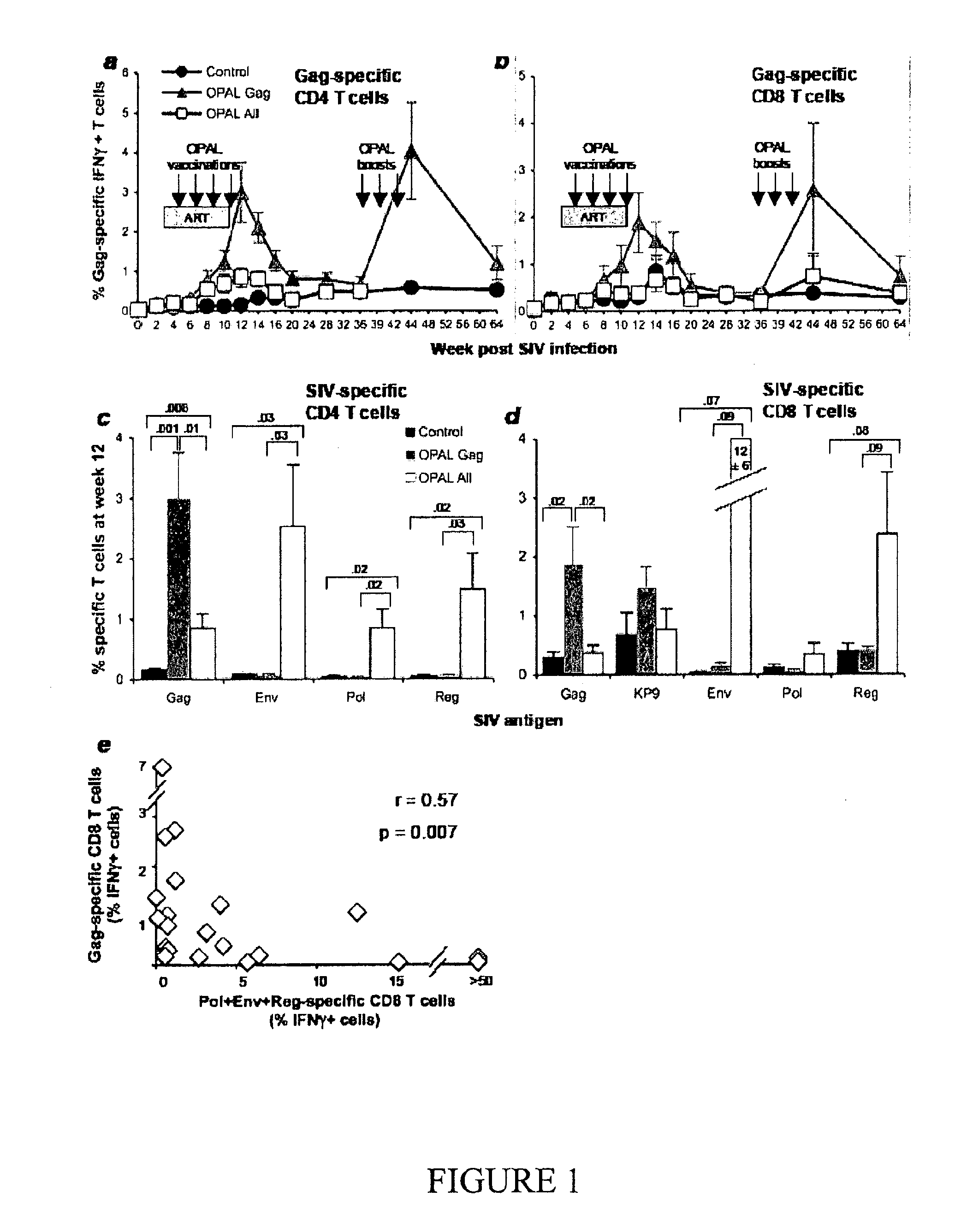
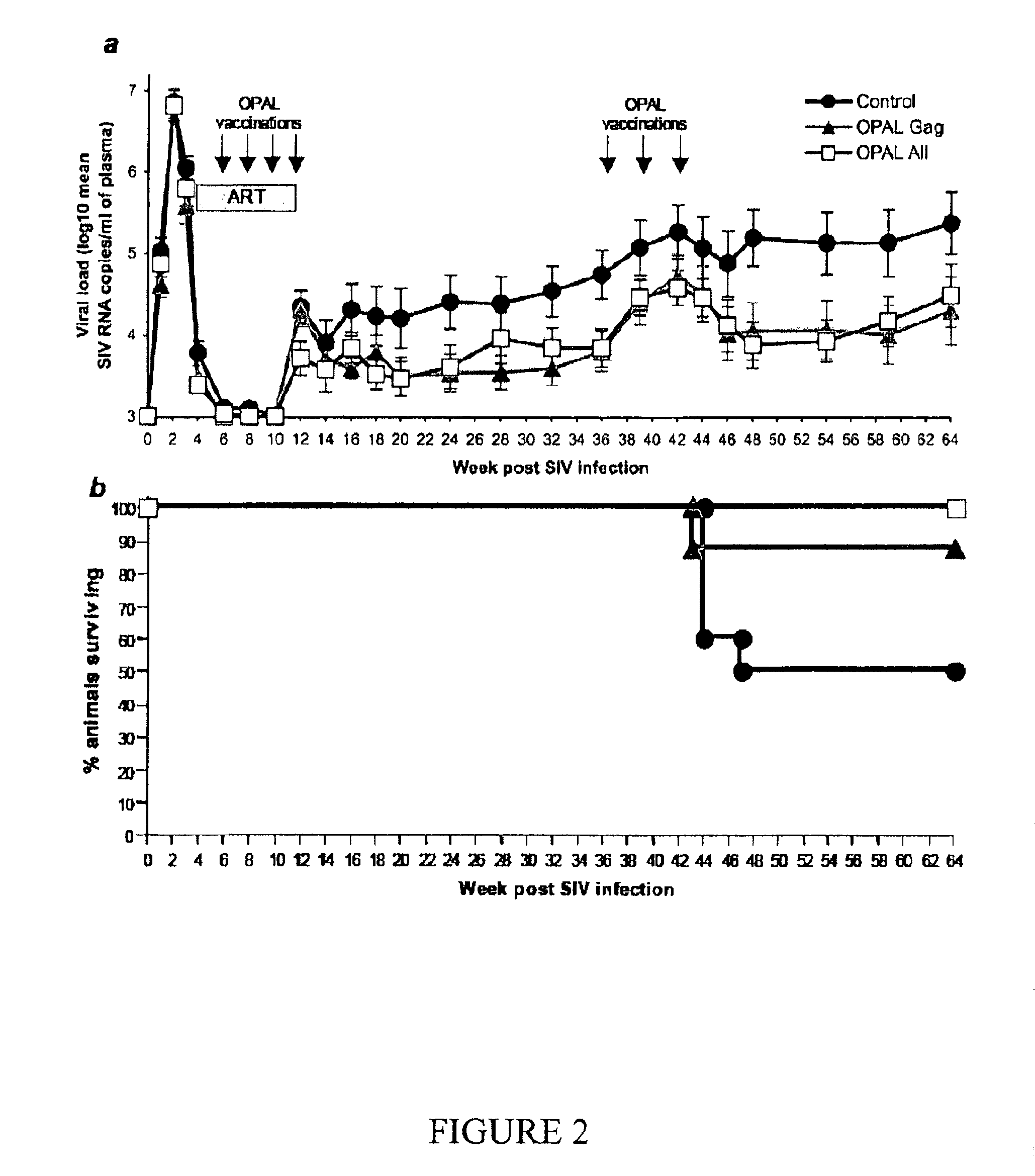
This invention discloses compositions that consist essentially of a Gag polypeptide or at least one portion thereof, and optionally antigen-presenting cells or their precursors, for treating or preventing lentiviral infections including the treatment or prevention of related acquired immunodeficiency diseases. In certain embodiments, the compositions consist essentially of a plurality of overlapping and / or non-overlapping peptides derived from a single Gag polypeptide or from different Gag polypeptides.
Owner:OPAL THERAPEUTICS
Lentiviral vector and application thereof in construction of immortalized cells
InactiveCN109295103AReduce cancer riskControlling the Immortalization ProcessGenetically modified cellsTransferasesPlasmid VectorInducer
The invention relates to the technical field of genetic engineering, in particular to a lentiviral vector and application thereof in construction of immortalized cells. According to the invention, bymeans of lentivirus infection, different immortalized key genes are selectively and targetedly integrated into a target cell genome, thus not only improving the immortalization efficiency, but also generating no harmful by-product in the cells. Because of simultaneous introduction of two immortalized genes, the plasmid vector has expanded application scope, can be applicable to more cell types, also the two groups of genes have different mechanisms in inducing immortalization, can complement each other to improve the immortalization efficiency. By adding an inducible promoter, TERT expressioncan be controlled by an inducer, the cell canceration probability is reduced, and the immortalization process of cells can be better controlled.
Owner:湖南丰晖生物科技有限公司
Bone marrow derived mesenchymal stem cell line for stable expression of exogenous EX-4 gene
InactiveCN107502594APromote proliferationInhibit apoptosisPancreatic cellsCulture processEnzyme digestionT cell
The invention provides an MSCs (mesenchymal stem cells) line for stable expression of EX-4. The MSCs line for stable expression of EX-4 is obtained by lentivirus infection. The lentivirus used by the invention is obtained by transfection of 293T cells with a PLVTH-pol-IRES-puro lentivirus overexpression system, and the lentivirus overexpression system comprises PLVTH-EX-4 expression plasmid, coating plasmid pMD2.G, and packaging plasmid p8.91. The PLVTH-EX-4 is obtained by double enzyme digestion of the lentivirus overexpression vector plasmid PLVTH-pol-IRES-puro and EX-4 target gene and then connection to T4 ligase. The cell line obtained by the invention on the one hand can enhance the anti anti-apoptosis ability, multiplication capacity and migration ability of MSCs itself, and on the other hand the EX-4 overexpressed MSCs group has better effects of inhibiting apoptosis and promoting proliferation on pancreatic beta cells than the MSCs group. Through the two aspects of effects, the cell line provided by the invention can recover the functions of pancreatic beta cells to a normal level, and improve the disadvantages of MSCs in treatment of diabetes.
Owner:CHINA PHARM UNIV
Methods and compositions for protection against lentiviral infections
ActiveUS20160340405A1Effective and broad protectionPeptide/protein ingredientsAntibody mimetics/scaffoldsViral infectionLentivirus Infections
The present invention provides methods and compositions for optimally co-expressing in a primate subject a tyrosylprotein sulfotransferase (TPST) and a lentiviral gp120-binding molecule to provide potent and long term protection against lentiviral infections.
Owner:UNIV OF FLORIDA RES FOUNDATION INC
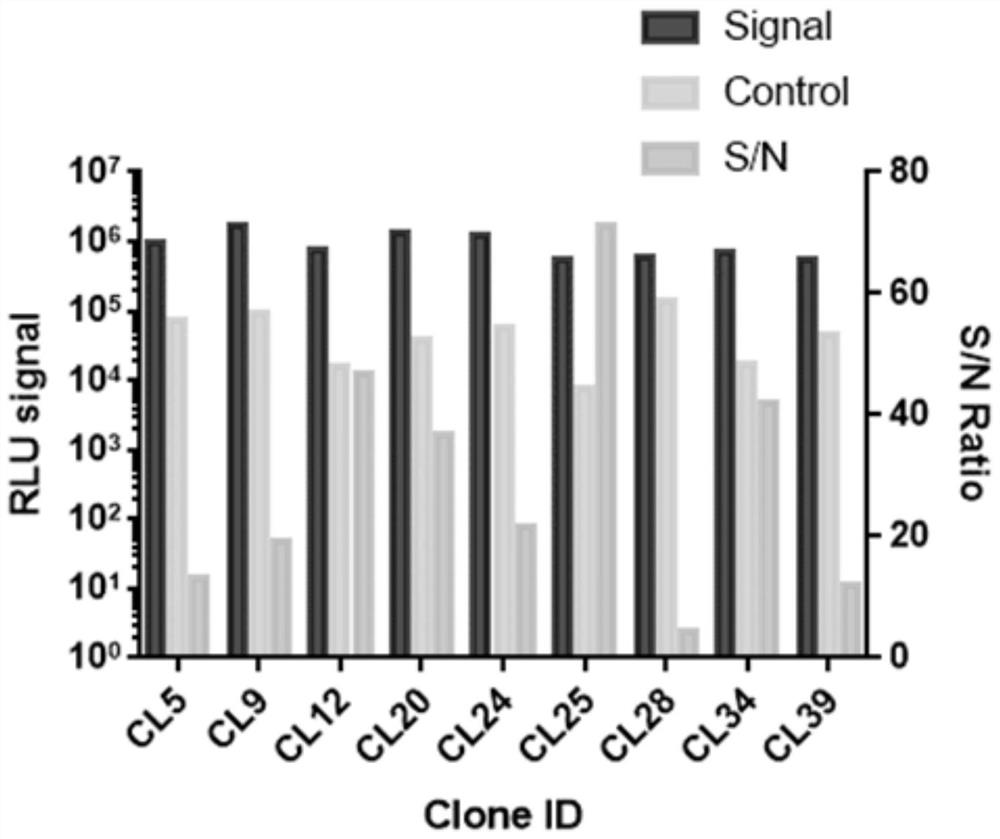
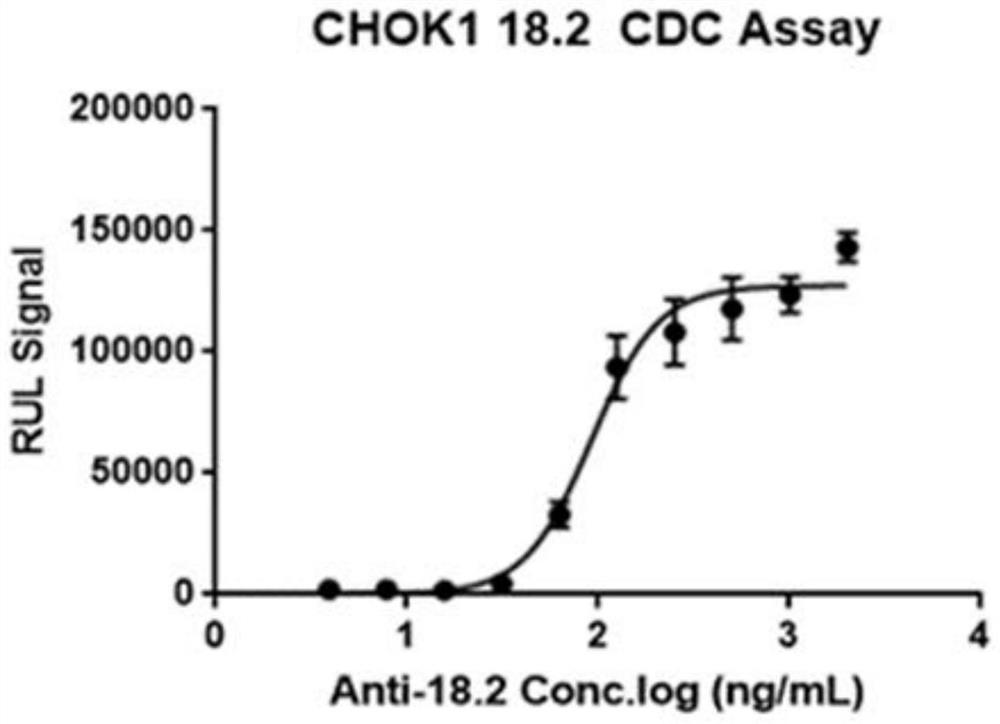
Construction and application of Claudin18.2 reporter gene CHO-K1 stably transfected cell strain
The invention discloses construction of a Claudin18.2 reporter gene CHO-K1 stably transfected cell strain, which comprises the following steps: transfecting CHO-K1 cells by using a lentivirus containing a Claudin18.2 gene, adding eukaryotic antibiotics for screening and monoclonal selection, and carrying out expression abundance detection and selection to obtain an appropriate high-expression Claudin18.2 stably transfected cell strain monoclonal; a Claudin18.2 stably transfected cell strain monoclonal antibody is infected by lentivirus containing a CMV-luciferase gene, and a proper Claudin18.2 reporter gene stably transfected cell strain is obtained by adding eukaryotic antibiotics for screening and monoclonal selection and performing functional evaluation. The invention also relates to a detection method for CDC killing by using the reporter gene stably transfected cell strain, a detection method for ADCC killing by using the reporter gene stably transfected cell strain, and the like. According to the present invention, the Claudin18.2 reporter gene CHO-K1 stably transfected cell strain is used for the CDC and ADCC cell killing determination method, such that the sensitivity is high, the external interference is not easily generated, and the stability of the used method is good.
Owner:宁波熙宁检测技术有限公司 +1
Evaluation method for therapeutic effect of CAR-T immune cells and device thereof
ActiveCN110129405AControllable and uniform sizePrediction is accurateMicrobiological testing/measurementDiagnosticsMatrigelTherapeutic effect
The invention relates to an evaluation method for therapeutic effect of CAR-T immune cells and a device thereof. Specifically, the method comprises the steps of: 1) preparation of CAR-T cell: constructing a CAR-T plasmid and packaging to lentivirus; obtaining a T cell of the subject, amplifying the cell after activation; and infecting the activated T cell with lentivirus, obtaining the CAR-T cellexpressing an antibody of the target gene; 2) preparation of 3D tumor-like organs: obtaining tumor tissue cells of the same subject as in step 1), obtaining primary cells, mixing primary cells with matrigel, and introducing matrigel and fluorocarbon oil containing primary cells into a three-way device respectively, and obtaining an organ-like sphere and culturing the material; 3) co-culturing the3D tumor-like organs with the CAR-T cells obtained in the step 1); and 4) measuring the survival rate of the tumor cells in the tumor-like organ spheres, and obtaining the therapeutic effect of the CAR-T cells. The method of the invention is simple, the preparation is fast, and the degree of reduction is high.
Owner:SHENZHEN GRADUATE SCHOOL TSINGHUA UNIV
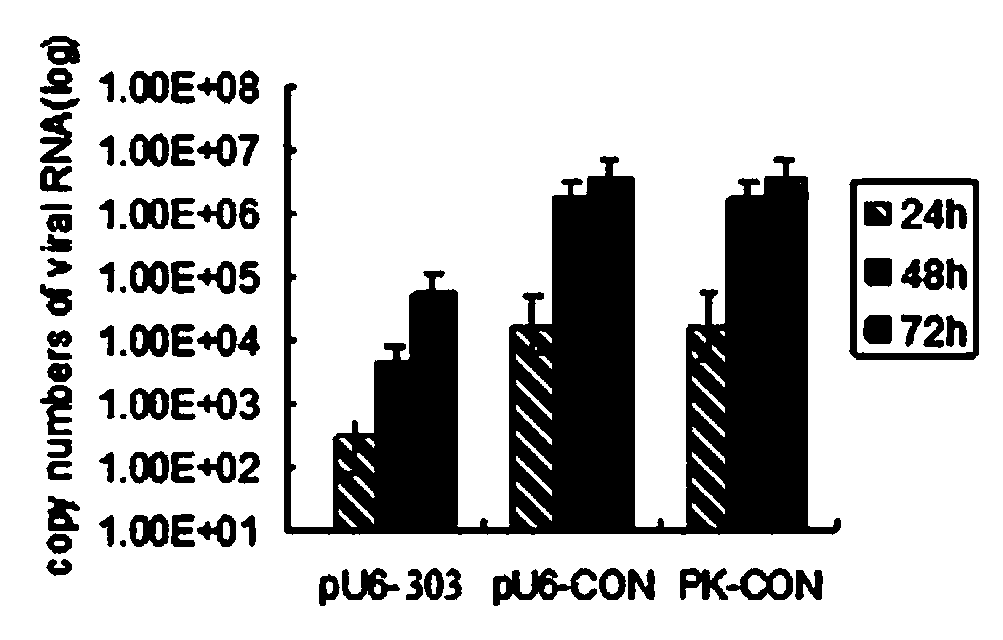
NS5B303shRNA (Short Hairclip Ribonucleic Acid) for inhibiting hog cholera virus replication and preparation method of NS5B303shRNA
The invention discloses an NS5B303shRNA (Short Hairclip Ribonucleic Acid) for inhibiting hog cholera virus replication. The NS5B303shRNA contains a siRNA (Small Interfering Ribonucleic Acid) sequence and is prepared through constructing a lentiviral expression vector of a shRNA, obtaining replication deficient lentivirus and lentivirus infection PK-15 cells (pig kidney cells) and verifying a hog cholera virus replication inhibiting effect. The sequence has a remarkable effect for inhibiting the replication of hop cholera viruses on sensitive cells. Through research on RNA interference to replication inside / outside the hog cholera viruses, a lentiviral vector mediated and stably-integrated RNA interfering technology for targeting a specific conserved gene fragment of a hog cholera virus genome is constructed, and siRNA transgenic animals for targeting the siRNA of hop cholera viruses are expected to be constructed. The necessary experimental data are accumulated for researching gene functions of the shRNA applied to the hop cholera viruses and preventing and treating hog cholera, and the previous preparation is provided for the animal breeding for disease resistance.
Owner:LANZHOU INST OF VETERINARY SCI CHINESE ACAD OF AGRI SCI
Methods for treating lentivirus infections
The present invention provides methods of identifying an agent that inhibits an activity of a lentiviral Vif protein. The present invention provides methods of identifying an agent that increases the level of active APOBEC3G in a cell. The present invention provides agents identified by a subject screening method; and further provides methods for treating lentivirus infections.
Owner:THE J DAVID GLADSTONE INST A TESTAMENTARY TRUST ESTABLISHED UNDER THE WILL OF J DAVID GLADS
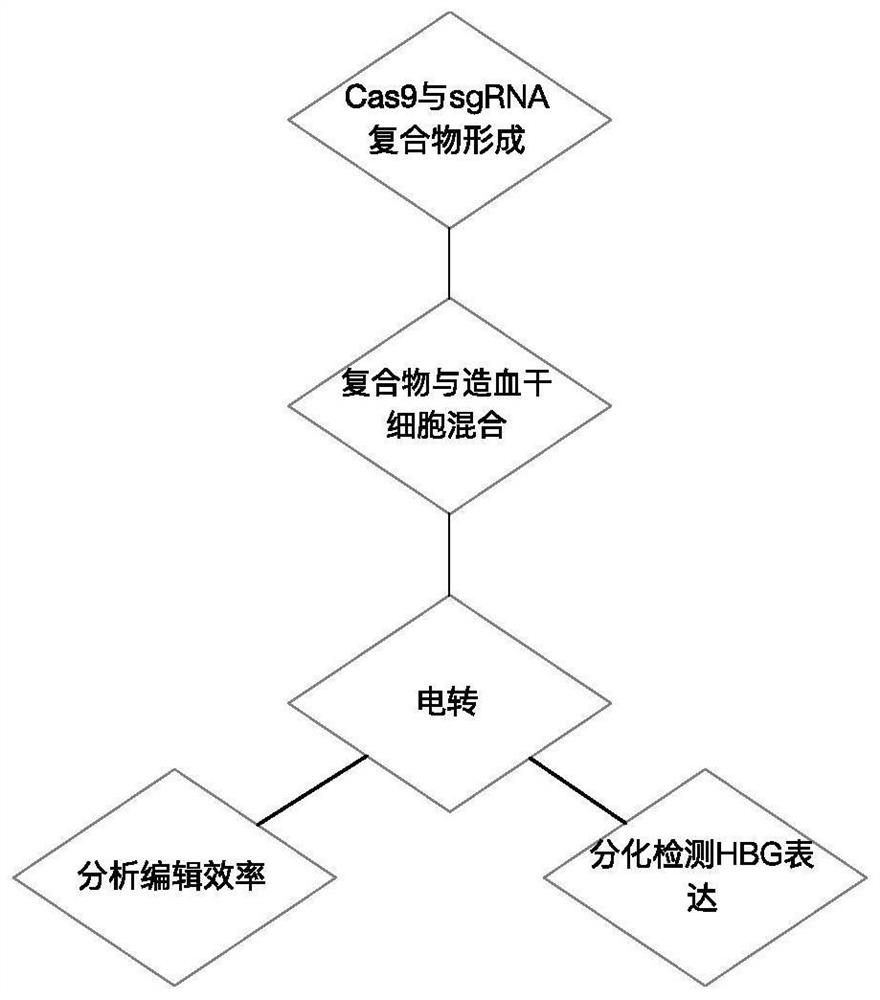
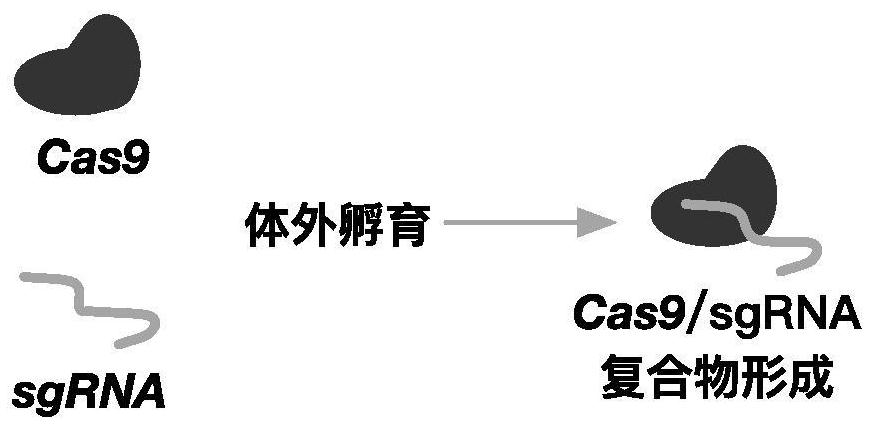
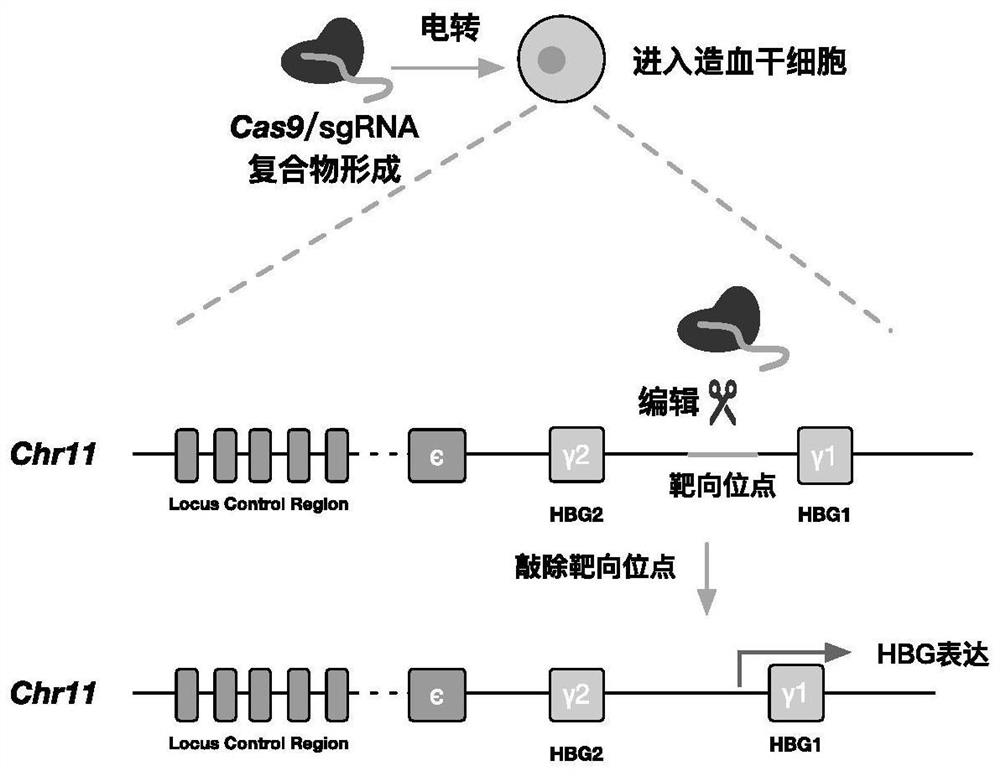
Application of CRISPR (clustered regularly interspaced short palindromic repeats) gene editing technology in treatment of thalassemia
PendingCN112011576AHigh clinical application prospectsSpecial deliveryHydrolasesGenome editingThalassemia
The invention provides application of a CRISPR (clustered regularly interspaced short palindromic repeats) gene editing technology in treatment of thalassemia. A CRISPR system in the application comprises at least one nuclease and at least one sgRNA, the sgRNA can target an inhibiting element of an HBG (gamma globin gene) promoter; the nuclease can cut the inhibiting element of the HBG promoter, the sgRNA comprises a targeting sequence targeting the inhibiting element of the HBG promoter, and the targeting sequence comprises a sequence as shown in SEQ ID No.1 or a complementary sequence as shown in SEQ ID No.1. Transferring a compound of the nuclease and the sgRNA into a cell by an electrotransformation method can knock out the inhibiting element on the HBG (gamma globin gene) promoter, and the relative content of HBG mRNA in cells is improved, and compared with a traditional lentiviral infection technology, exogenous DNA random integration caused by viruses is avoided, and the prospect of clinical application is improved.
Owner:EAST CHINA NORMAL UNIV +1
Methods of identifying agents for inhibiting lentivirus replication
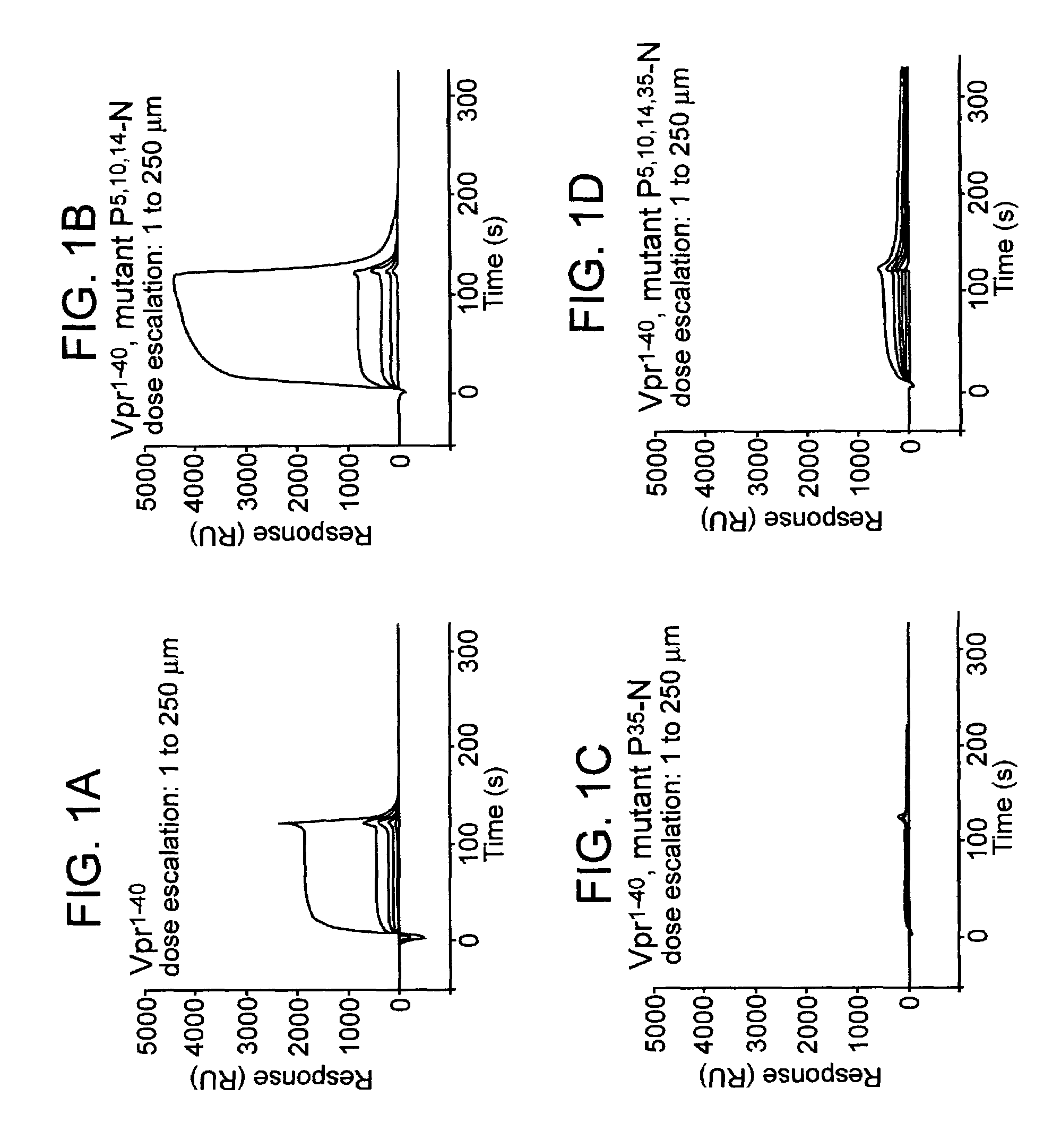
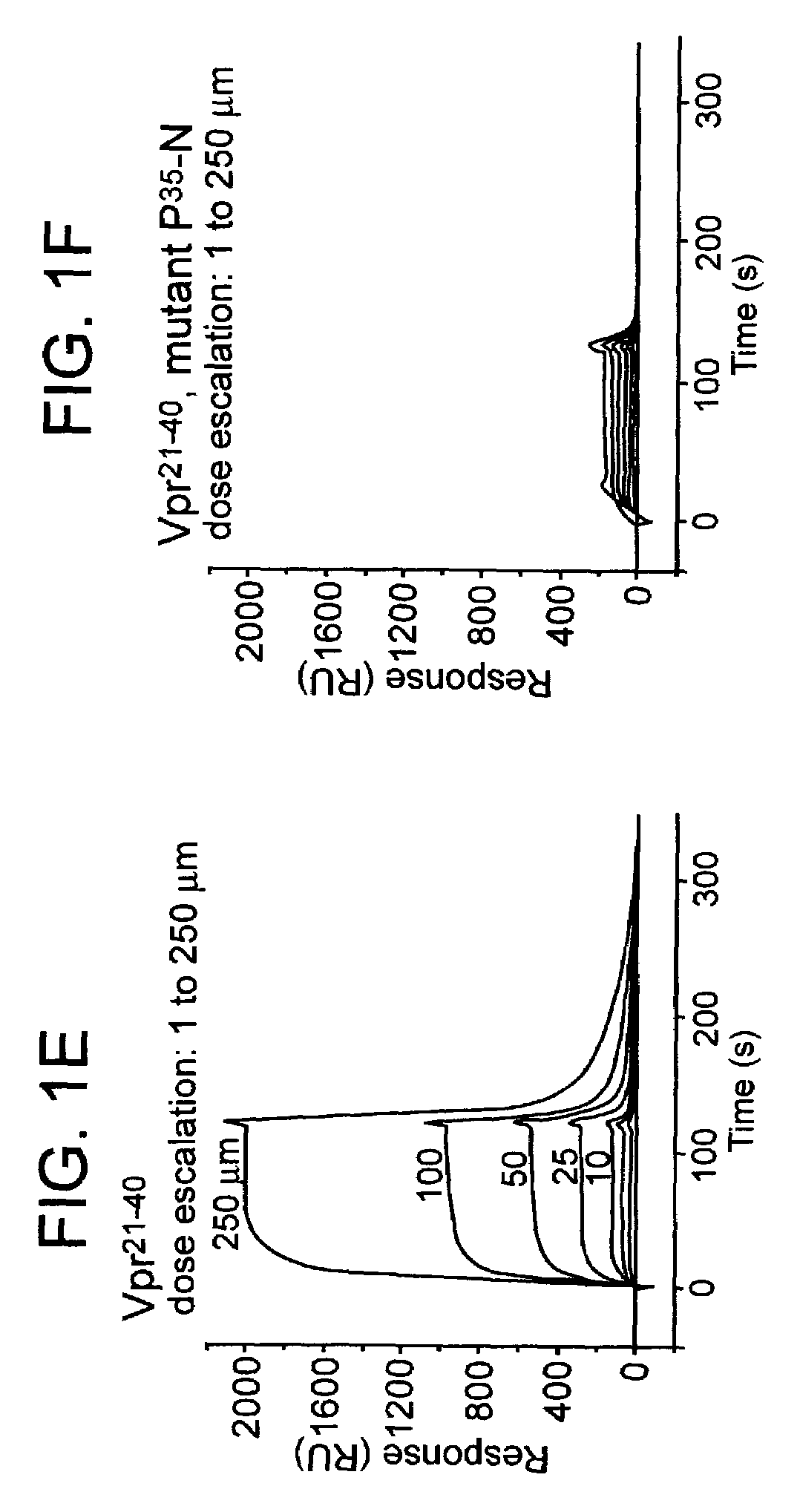
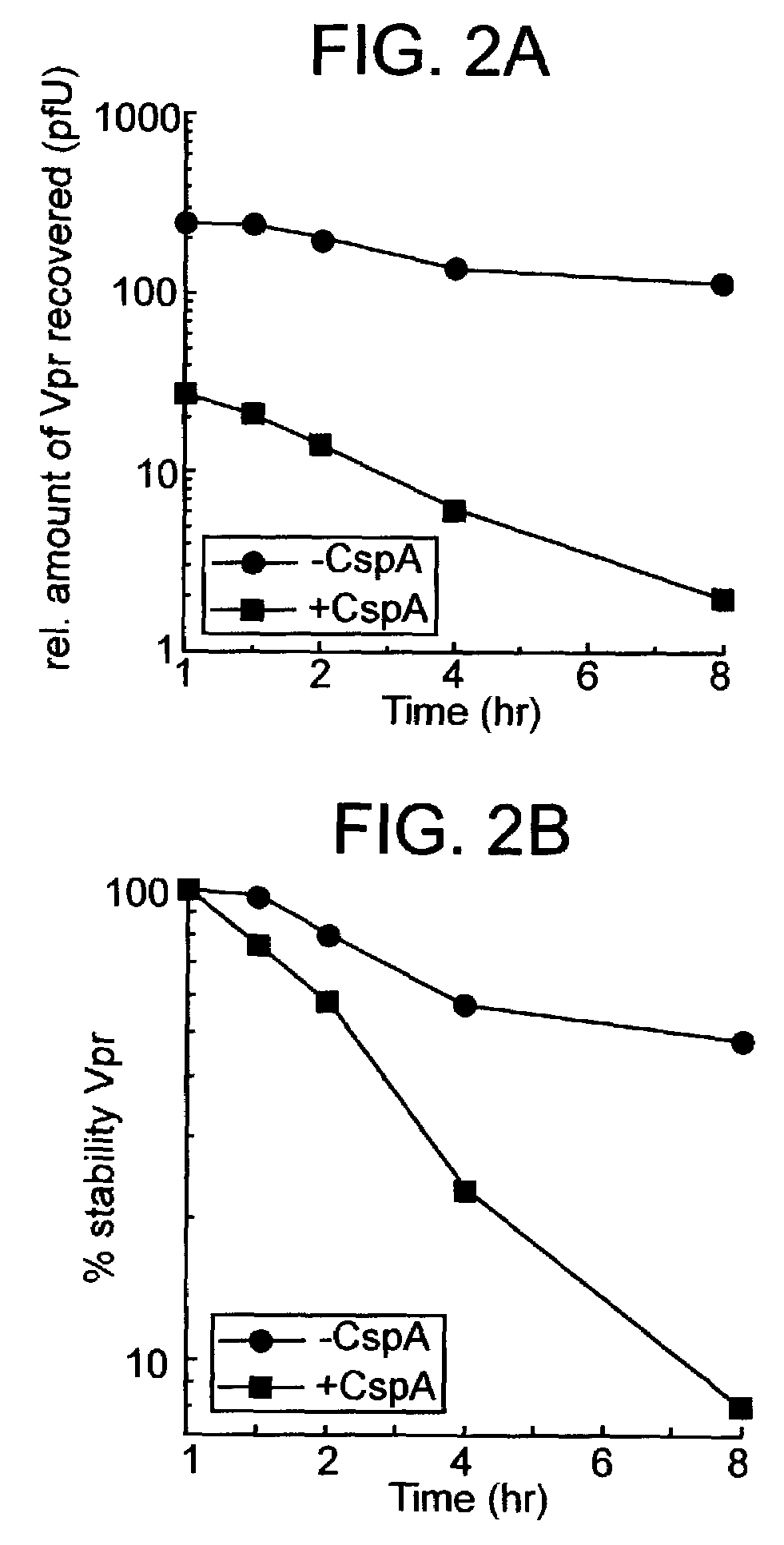
InactiveUS7108988B2Inhibitory activityInduce lossCompound screeningApoptosis detectionVpr ProteinScreening method
The present invention provides screening methods for identifying a compound that induces loss of the lentiviral protein Vpr; screening methods for identifying compounds that inhibit the peptidyl-prolyl cis / trans isomerase (PPIase) activity of a protein that catalyzes cis-trans isomerization of cis-peptidylprolyl bonds in Vpr; and compounds identified by the screening methods. The compounds are useful for treating a lentiviral infection. The present invention further provides methods of inducing loss of the lentiviral protein Vpr; methods of inhibiting lentivirus viral replication; and methods of treating a lentivirus infection in an individual. The methods generally involve administering to an individual infected with the lentivirus an effective amount of a compound that induces Vpr loss and / or that inhibits PPIase activity of a protein that catalyzes cis-trans isomerization of cis-peptidylprolyl bonds in Vpr.
Owner:THE J DAVID GLADSTONE INST A TESTAMENTARY TRUST ESTABLISHED UNDER THE WILL OF J DAVID GLADS
Macrophage derived chemokine (MDC) as an anti-viral agent for the treatment and prevention of lentivirus infection
InactiveUS6548631B1Reduce viral loadPeptide/protein ingredientsGenetic material ingredientsMacrophage-Derived ChemokineImmunodeficiency virus
The present invention relates to therapeutic compositions of macrophage derived chemokine (MDC) and methods for treating and preventing infection by a lentivirus, particularly an immunodeficiency virus, particularly HIV, using MDC proteins, nucleic acids and / or derivatives or analogues thereof. The present invention further relates to methods for detection and prognosis of lenivirus infection, particularly HIV infection using MDC as a prognostic indicator. The present invention further provides MDC proteins, nucleic acids encoding such proteins, that have amino-terminal sequences that differ from other MDC isolates. Recombinant host cells and methods of production are also provided.
Owner:AKZO NOBEL NV +1
Sequencing library construction method for detecting lentivirus insertion sites, and lentivirus insertion site detection method
PendingCN113046835AEasy to analyzeEase of evaluationMicrobiological testing/measurementLibrary creationInfected cellLentivirus Infections
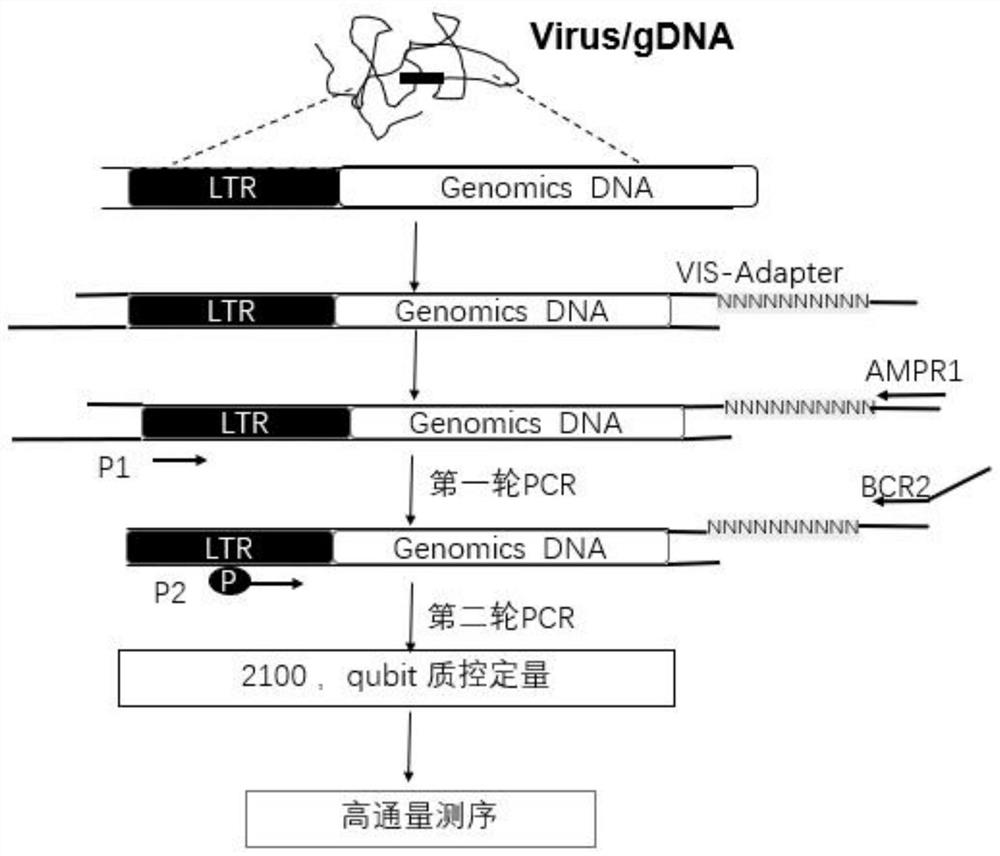
The invention relates to a sequencing library construction method for detecting lentivirus insertion sites, and a lentivirus insertion site detection method. The sequencing library construction method comprises the following steps: extracting genome DNA from lentivirus infected cells; carrying out fragmentation on the genome DNA and processing the genome DNA into a form suitable for linker connection; connecting asymmetric double-chain connectors to two ends of the fragmented genome DNA, wherein the asymmetric double-chain connectors comprise a long-chain sequence and a short-chain sequence; carrying out first-round PCR amplification on a joint connection product; performing second-round PCR amplification on a product of the first-round PCR amplification; and cyclizing the product of the second-round PCR amplification to obtain a cyclized sequencing library suitable for computer sequencing. The methods have the characteristics of simplicity in operation, short experiment time, low cost, small initial quantity, high flux, multiple analyzable aspects and the like, and can better perform accurate analysis and evaluation on the virus insertion site in gene therapy.
Owner:深圳市禾沐基因生物技术有限责任公司
Method of establishing hepatitis B virus infection cell model by using porcine primary hepatocyte and hNTCP recombinant lentivirus
ActiveCN107988260AIncreased susceptibilityUnrestricted sourceCell dissociation methodsArtificial cell constructsLiver tissueSingle cell suspension
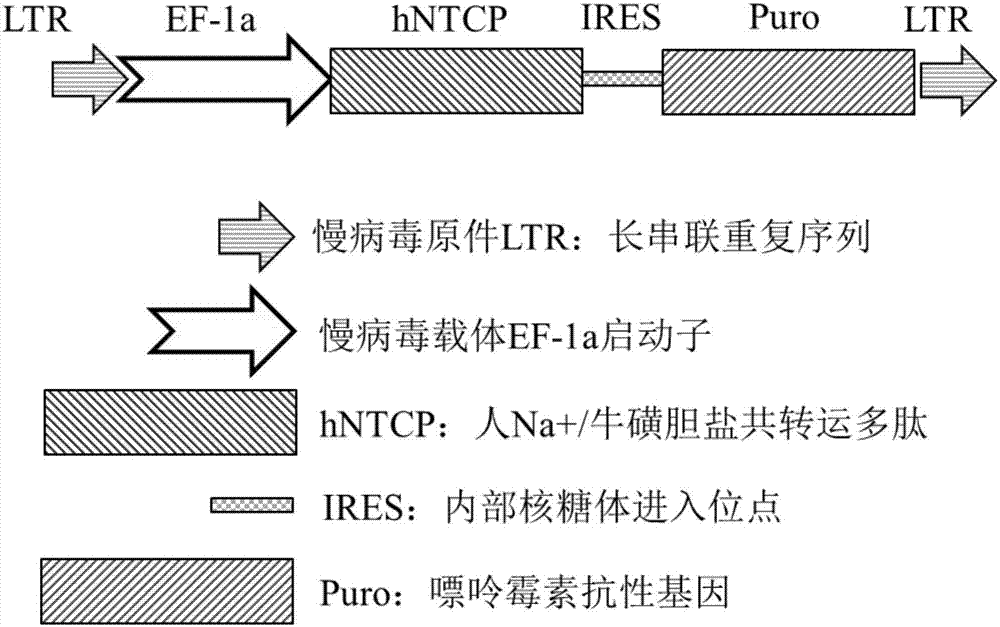
The invention discloses a method of establishing a hepatitis B virus infection cell model by using porcine primary hepatocyte and hNTCP recombinant lentivirus and belongs to the technical field of cell modification. The method comprises the following steps: S1, preparing an hNTCP recombinant lentivirus concentrated liquid; S2, preparing liver tissues which are fully digested; S3, preparing the porcine primary hepatocyte; S4, preparing a porcine primary hepatocyte single cell suspension; S5, preparing a cell suspension for paving a plate; S6, paving cell plate and culturing; S7, carrying out hNTCP recombinant lentivirus infection on the porcine primary hepatocyte; and S8, establishing the hepatitis B virus infection cell model. According to the invention, the recombinant lentivirus containing an hNTCP gene is constructed in vitro, the hNTCP gene is integrated into the primary hepatocyte genome through the lentivirus with high efficiency of infection to achieve stable overexpression of hNTCP, so that the primary hepatocyte supports hepatitis B virus infection.
Owner:立沃生物科技(深圳)有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com