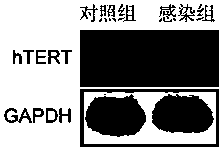
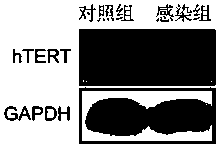
Lentiviral vector and application thereof in construction of immortalized cells
A technology of immortalization and lentivirus, applied in the field of genetic engineering, can solve problems such as complex mechanism, decay, and difficulty in control, and achieve the effects of expanding the application range, reducing the possibility of canceration, and improving the efficiency of immortalization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
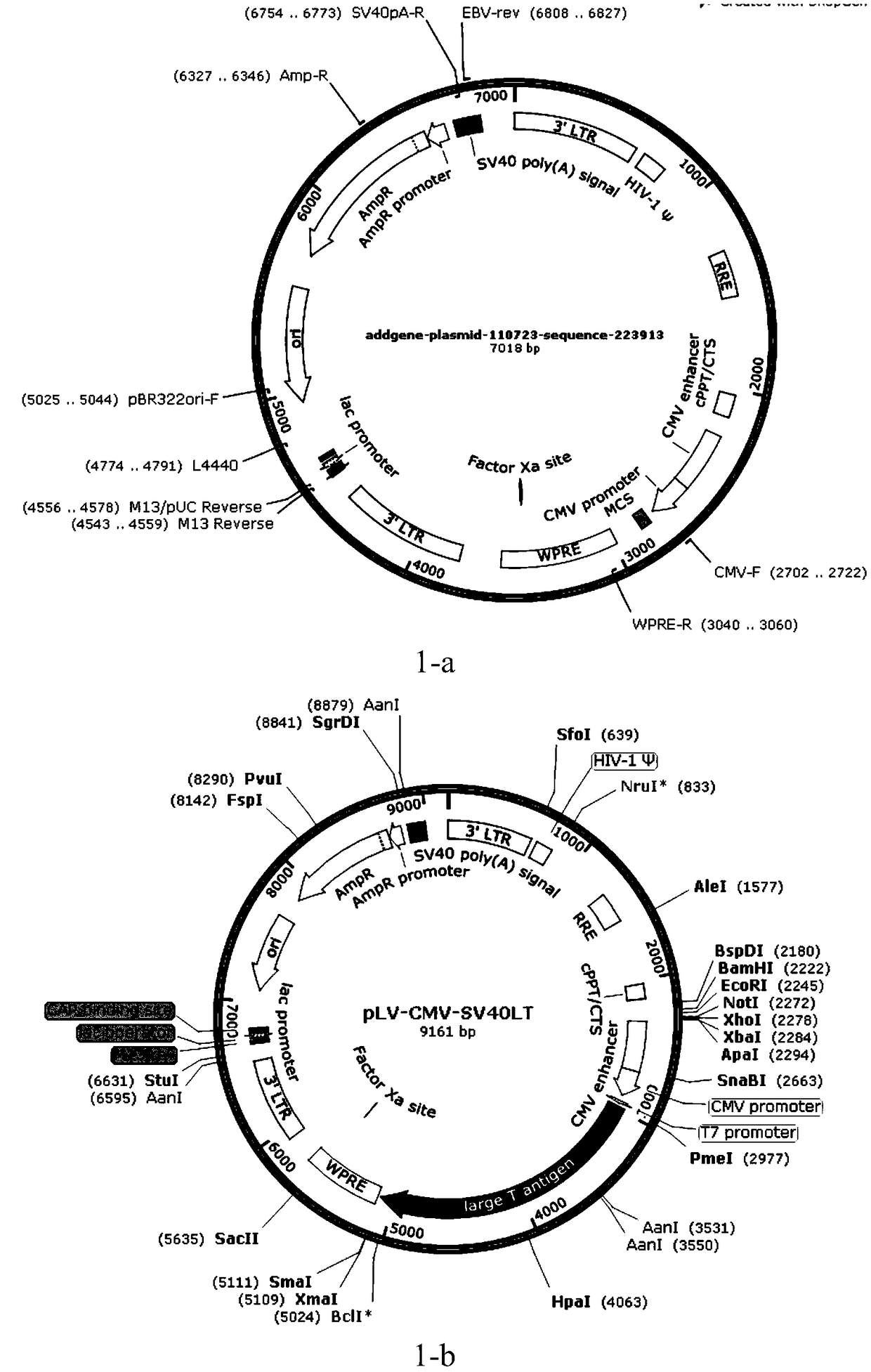
[0089] 1. Using the lentiviral vector pLV-CMV-SV40LT as the base expression vector, construct the pLV-CMV-HPV16E6 vector.
[0090] 2. Synthesize the HPV16E6 sequence (NC_001526.4, SEQ ID NO: 4), and add Pme I (GTTTAAAC) and Sma I (CCCGGG) restriction site sequences at both ends
[0091] 3. The pLV-CMV-SV40LT vector ( figure 1 ) in the SV40LT sequence is replaced by the HPV16E6 sequence to obtain the pLV-CMV-HPV16E6 vector ( figure 2 ).
[0092] 4. Synthesize the TRE3G sequence (SEQ ID NO: 2), and add BamH I (GGATCC) and EcoR I (GAATTC) restriction site sequences at both ends.
[0093] 5. Construct the TRE3G sequence on the pLV-CMV-HPV16E6 vector by double enzyme digestion to obtain the pLV-TRE3G-CMV-HPV16E6 vector.
[0094] 6. Synthesize hTERT sequence (NM_198253.2, SEQ ID NO: 1), and add EcoR I (GAATTC) and Xba I (TCTAGA) restriction site sequences at both ends.
[0095]7. Construct the hTERT sequence into pLV-TRE3G-CMV-HPV16E6 by means of double enzyme digestion to obta...
Embodiment 2
[0097] 1. Using the lentiviral vector pLV-CMV-SV40LT as the base expression vector, construct the pLV-TRE3G-hTERT-CMV-SV40LT vector.
[0098] 2. Synthesize the TRE3G sequence (SEQ ID NO: 2), and add BamH I (GGATCC) and EcoR I (GAATTC) restriction site sequences at both ends.
[0099] 3. The TRE3G sequence was constructed on the pLV-CMV-SV40LT vector by double enzyme digestion to obtain the pLV-TRE3G-CMV-SV40LT vector.
[0100] 4. Synthesize hTERT sequence (NM_198253.2, SEQ ID NO: 1), and add EcoR I (GAATTC) and Xba I (TCTAGA) restriction site sequences at both ends.
[0101] 5. The hTERT sequence was constructed into pLV-TRE3G-CMV-SV40LT by means of double enzyme digestion to obtain the pLV-TRE3G-hTERT-CMV-SV40LT vector ( image 3 ).
Embodiment 3
[0103] The pMD2.G vector and psPAX2 vector will be used to package the pLV-TRE3G-hTERT-CMV-HPV16E6 vector into a lentivirus. The packaging method is:
[0104] 1. Divide 293T cells into 7×10 6 Each dish was inoculated, and a total of 3 dishes were inoculated. Add DMEM medium containing 10% fetal bovine serum, place at 37°C, 5% CO 2 cultured in an incubator.
[0105] 2. After 24 hours, 10 μg pMD2.G, 12 μg psPAX2, and 10 μg pLV-TRE3G-hTERT-CMV-HPV16E6 plasmids were co-transfected into 293T cells.
[0106] 3. Collect the cell supernatant 48h and 72h after transfection, filter to remove impurities, concentrate to 1ml with an ultrafiltration tube, divide into 5 tubes, and store at -80°C.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com