Recombinant crassostrea gigas high-mobility family protein r-CgHMGB1, preparation method and application thereof
A high-mobility, r-cghmgb1 technology, applied in the field of molecular biology, can solve problems such as antibacterial and immune functions of high-mobility group proteins in oysters that have not been seen.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
experiment example 1
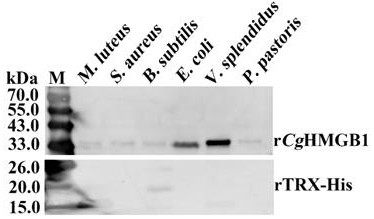
[0036] Experimental Example 1: Recombinant oyster high mobility group protein r- C g HMGB1 bacterial binding activity detection
[0037] Detection of recombinant long oyster high mobility group protein r- based on Western blotting C g HMGB1 against two Gram-negative bacteria: Vibrio splendidus ( Vibrio splendidus JZ6 , abbreviated as V. splendidus group, the same below) and Escherichia coli ( Escherichia coli , E. coli ), three Gram-positive bacteria: Micrococcus luteus ( Micrococcus luteus , M. luteus ), Staphylococcus aureus ( Staphylococcus aureus , S. aureus ) and Bacillus subtilis ( Bacillus subtilis , B. subtilis ) and a fungus Pichia pastoris ( Pichia pastoris GS115 , P. pastoris ) binding activity. The sources of the strains used are as follows: Vibrio resplendent was purchased from Beijing Microorganism Culture Collection Center, Escherichia coli was purchased from Beijing Quanshijin Company, Staphylococcus aureus was purchased from Beijing M...
experiment example 2
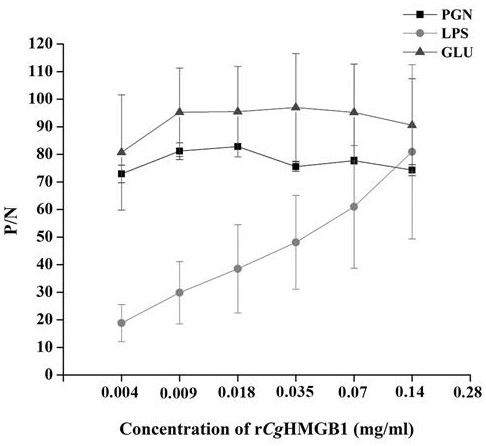
[0057] Experimental Example 2: Recombinant oyster high mobility group protein r- C g Carbohydrate binding activity assay of HMGB1
[0058] Detection of recombinant high mobility group protein r- C g Binding of HMGB1 to various PAMPs. The peptidoglycan (PGN), lipopolysaccharide (LPS) and glucan (GLU) used were purchased from Sigma.
[0059] The specific operation steps are as follows:
[0060] (1) Dissolve 10 μg of various PAMPs in Na 2 CO 3 with NaHCO 3 According to 15 mmol·L -1 and 35 mmol·L -1 Concentration configuration of the coating solution with a pH value of 7.6, 100 μL per well to coat the microtiter plate, and refrigerate overnight at 4 °C;
[0061] (2) Discard the coating liquid and wash 3 times with TBST, 5 min each time;
[0062] (3) After washing, add 200 μL of 3% BSA to the wells and seal them in a constant temperature incubator at 37°C for 1 h;
[0063] (4) Wash with TBST 3 times after sealing, 5 min each time;
[0064] (5) Add 100 μL of diluted conce...
experiment example 3
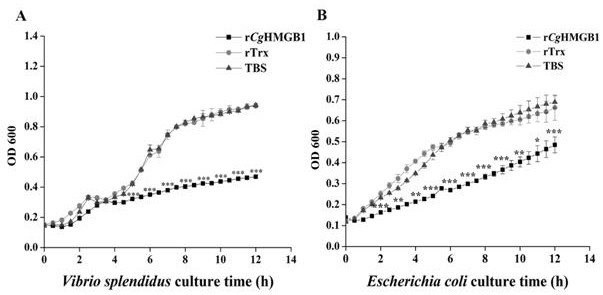
[0072] Experimental Example 3: Recombinant oyster high mobility group protein r- C g Bacteriostatic activity assay of HMGB1
[0073] Recombinant oyster high mobility group protein r- C g HMGB1 was incubated with two kinds of bacteria (Vibrio splendidus and Escherichia coli), and the concentration of the bacterial solution was detected using a microplate reader (Tecan Infinite M1000 PRO), and the recombinant long oyster high mobility group protein r- C g Bacteriostatic activity of HMGB1.
[0074] The strain source is as above.
[0075] The specific operation is as follows:
[0076] (1) This experiment is divided into three groups: the embodiment of the present invention recombinant long oyster high mobility group protein r- C g HMGB1 treatment group, rTrx control group (negative control), TBS control group (blank control), the number of repetitions of each experimental group is not less than three parallel;
[0077] (2) Cultivate the above two microorganisms separately and...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap