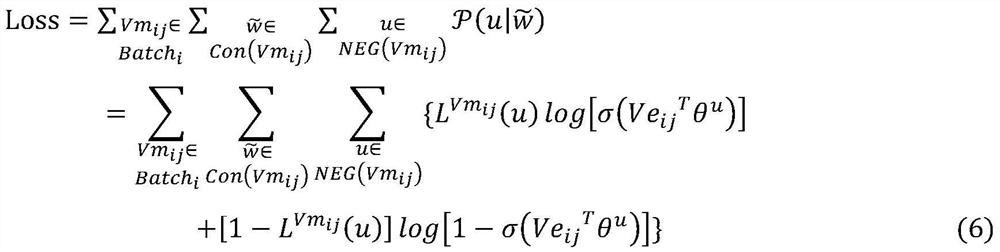Anti-HBV small molecule drug prediction model based on expression learning and construction method thereof
A technology for prediction models and construction methods, which can be used in chemical property prediction, molecular design, etc., and can solve problems such as delaying drug research and development.
Pending Publication Date: 2022-01-11
NANTONG UNIVERSITY
View PDF0 Cites 0 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
[0003] Limited by the routine drug development process, large-scale drug molecule design, synthesis, and anti-HBV activity experiments consume a lot of manpower, material resources, time and capital, which delays the speed of drug development
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment
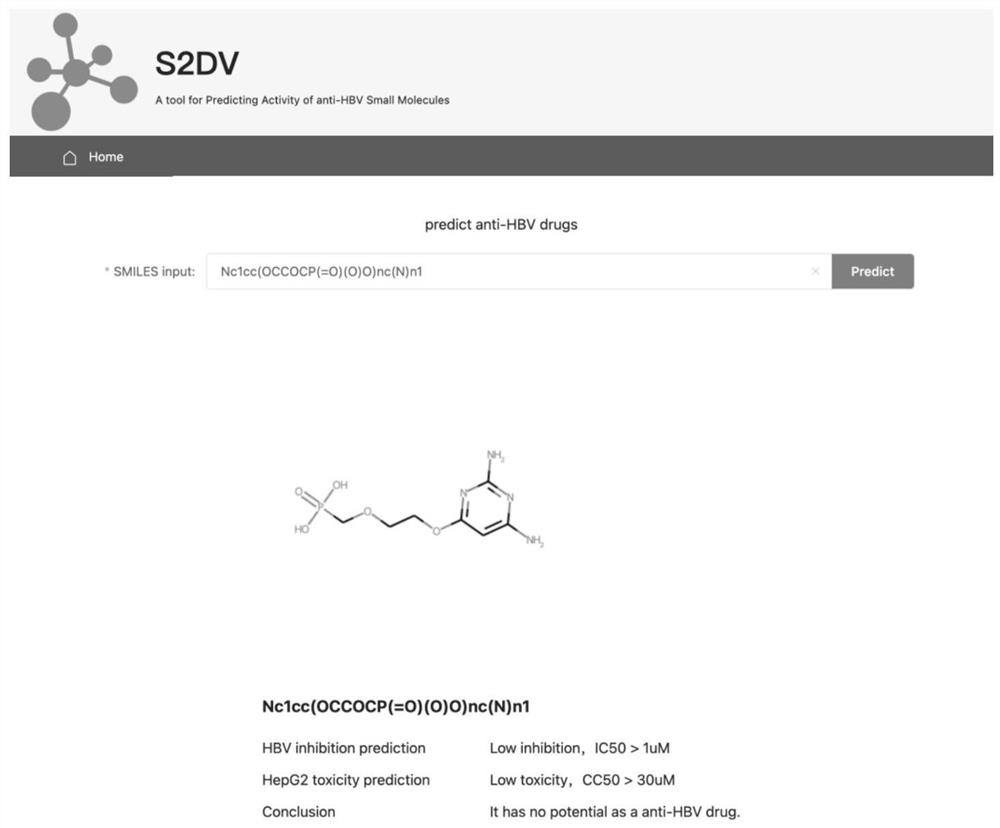
[0095] 1. Evaluation of anti-HBV activity and cytotoxicity: The in vitro anti-HBV activity and cytotoxicity of all compounds in HepG2 2.2.15 cells were evaluated by real-time quantitative PCR and MTT method, respectively. The concentration of compound required to inhibit DNA replication by 50% was defined as IC50 and the concentration of compound that induced 50% death of HepG2 2.2.15 cell cultures was defined as CC50.
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
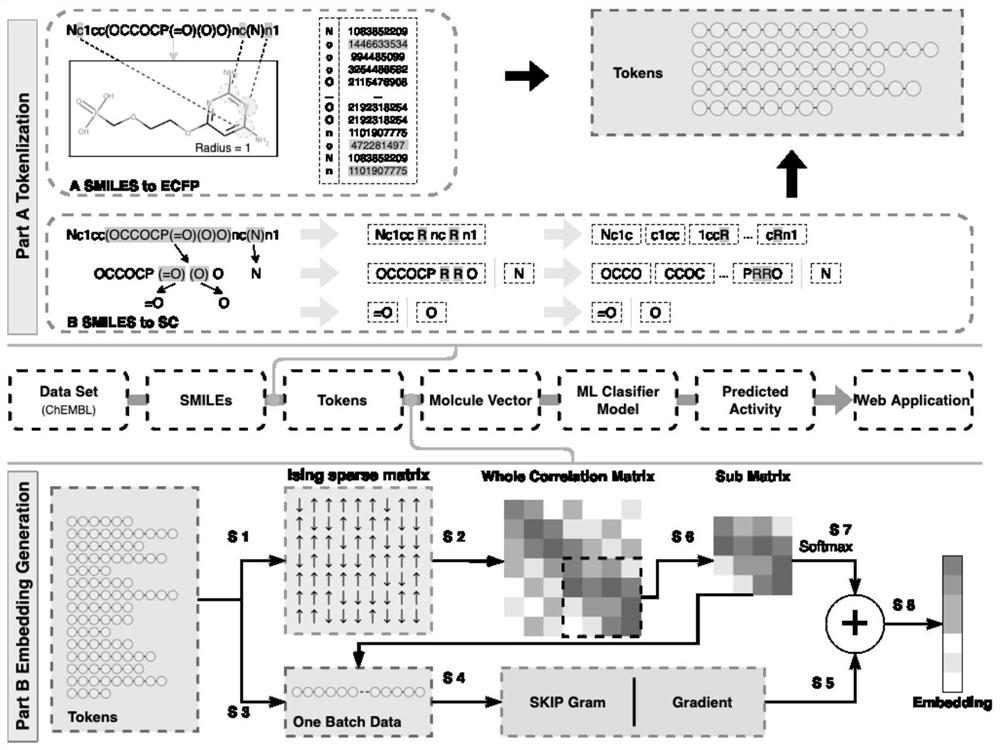
The invention provides an anti-HBV small molecule drug prediction model based on expression learning and a construction method thereof, relates to the technical field of drug research, and aims to perform expression learning on SMILES codes by using an NLP method through processing the SMILES codes. A machine learning method is used for a supervised classification model of SMILES coded space vectors, and the activity of small molecules to specific targets is predicted in a classified mode in the model. Compared with the prior art, the performance superior to that of traditional Word2vec is embodied on different target point data sets. Meanwhile, in downstream tasks after vector training, deterministic advantages are generated in the aspects of predicting the inhibition rate of the compound to HBV and the toxicity of the compound to hepatocytes, and the model has good capacity of screening potential anti-HBV drugs. The method can be widely applied to drug-likeness prediction of other different target compounds, so that the drug research and development process is simplified.
Description
technical field [0001] The invention relates to the technical field of drug research and development, in particular to a method for constructing a predictive model of anti-HBV small molecule drugs based on representation learning. Background technique [0002] In the process of drug development, designing and synthesizing compounds with high activity against specific targets is the primary task of medicinal chemistry. Usual drug design relies more on various types of high-throughput virtual screening, fragment or structure-based drug design and the experience of drug developers, and lacks a convenient, quick and accurate way to evaluate the pharmacological activity of drug molecules against specific targets. [0003] Limited by the routine drug development process, large-scale drug molecule design, synthesis, and anti-HBV activity experiments consume a lot of manpower, material resources, time, and capital costs, delaying the speed of drug development. Through the artificia...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More IPC IPC(8): G16C20/50G16C20/30
CPCG16C20/50G16C20/30
Inventor 王理邵劲松尹泽宇潘文洁
Owner NANTONG UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com