Detection of colorectal cancer
A technology for colorectal cancer and subjects, applied in the field of colorectal cancer treatment, colorectal cancer screening, and colorectal cancer screening, can solve the problems of insufficient colorectal cancer screening technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0253] Example 1. Identification of methylation biomarkers associated with colorectal cancer
[0254] This example includes the identification of one or more CpG loci that are hypermethylated in colon and rectal cancer compared to healthy controls. In particular, the experiments of this example examined CpG methylation in samples from (i) colon cancer in 341 subjects previously diagnosed with colon cancer who had not previously received chemotherapy therapy or radiotherapy; (ii) rectal cancer in 118 subjects previously diagnosed with rectal cancer who had not previously received chemotherapy or radiotherapy; (iii) 40 subjects not diagnosed with rectal cancer Colon of healthy control subjects with colorectal cancer; (iv) leukocytes of 10 healthy control subjects not diagnosed with colorectal cancer. Tissue samples were fresh frozen tissue.
[0255]Samples were analyzed for DNA methylation by a global methylomics analysis platform (Infinium HumanMethylation450 (HM450) bead tes...
Embodiment 2
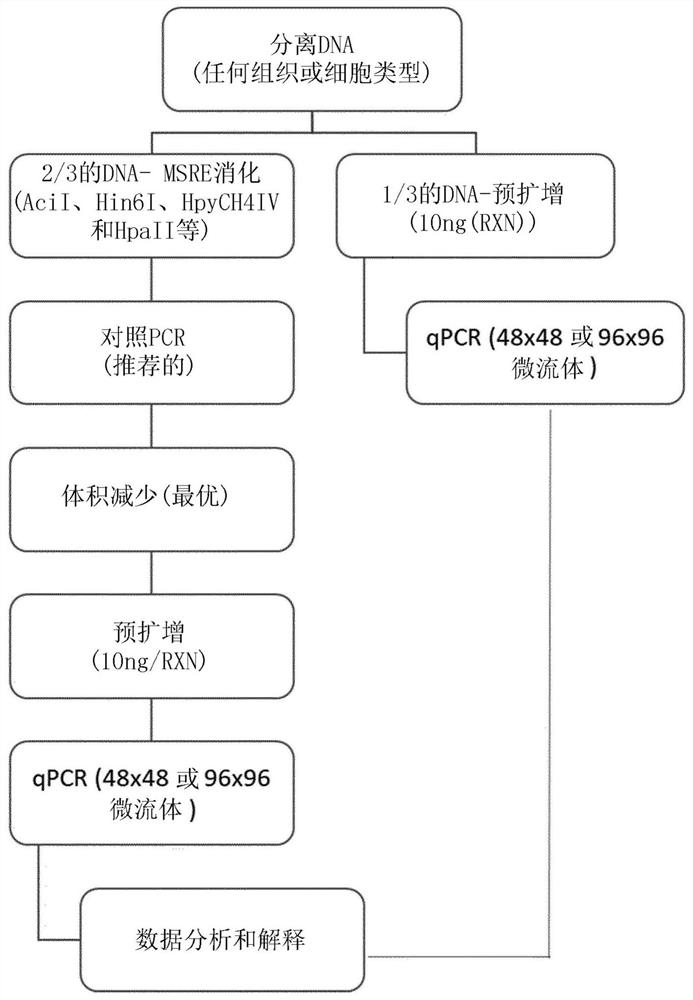
[0258] Example 2: Development of a cell-free DNA assay for methylation biomarkers by MSRE-qPCR
[0259] This example develops a circulating cell-free DNA (cfDNA)-based assay to determine the methylation status of colorectal cancer methylation biomarkers. cfDNA is incomplete and fragmented, and the mechanism by which cfDNA is transported from cancer cells to the blood (as part of it called circulating tumor DNA) is unknown. At least because the 253 methylation biomarkers of Example 1 were identified from tissue samples, it was not known prior to the experiments of this Example whether the identified colorectal cancer methylation biomarkers could be adequately analyzed from cfDNA to Successful capture of ctDNA fractions allows identification of subjects or samples of colorectal cancer.
[0260] As a critical step in determining whether the colorectal cancer methylation biomarkers identified in Example 1 can be adequately analyzed from cfDNA to successfully capture the fraction ...
Embodiment 3
[0266] Example 3: MSRE-qPCR of cfDNA successfully differentiated subjects by colorectal cancer status
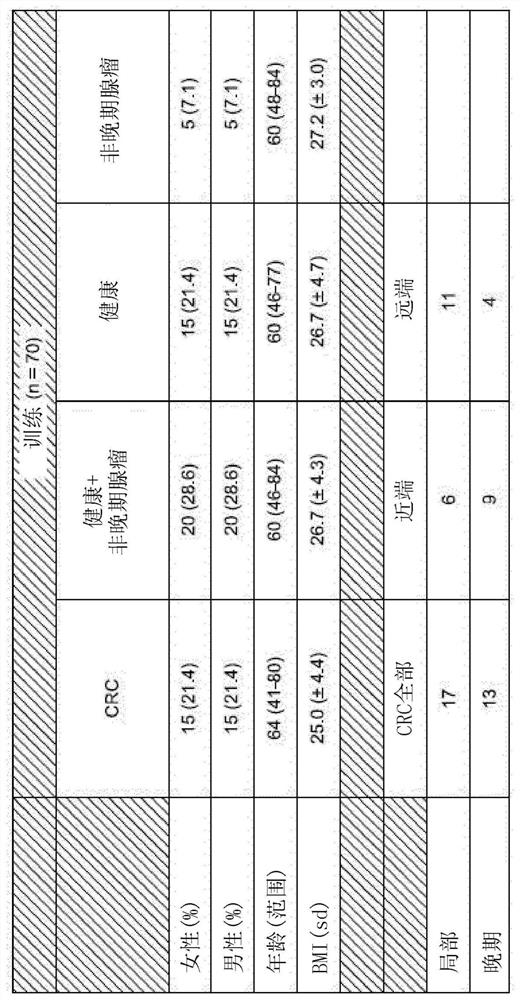
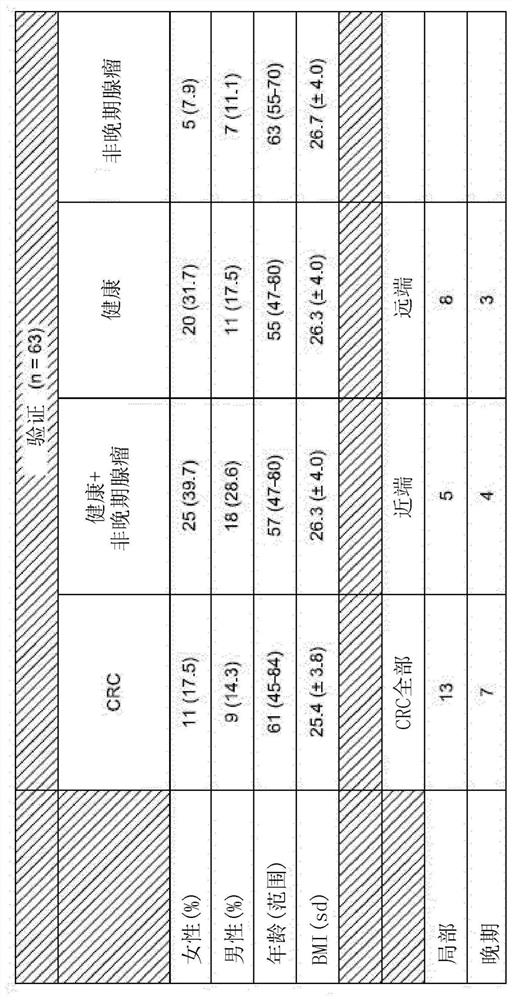
[0267] To probe the clinical diagnostic and prognostic capabilities of the identified methylated biomarkers, MSRE-qPCR oligonucleotides covering 180 methylated biomarker sites were tested in cfDNA extracted from the plasma of human subjects Acid primer pairs amplify the DMR, and appropriate controls. Subjects were undiagnosed individuals seeking or receiving a diagnosis of probable colorectal cancer, allowing methylation biomarker analysis to be performed prior to conventional colorectal cancer diagnostic testing and then compared to subsequent conventional diagnoses. In particular, cfDNA was obtained from undiagnosed individuals seeking or being obtained at screening centers and oncology clinics in Spain and the United States between 2017 and 2018 for a possible colorectal cancer diagnosis. The first subject group included 70 such individuals (see figure 2 description of...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com