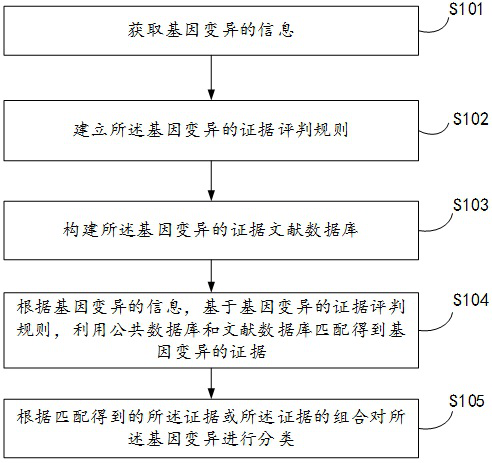
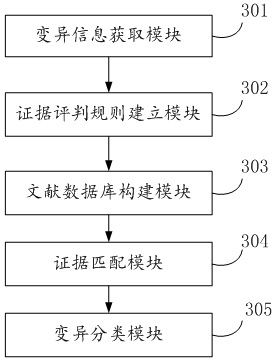
An automatic classification method, device and electronic device for gene variation
A gene variation and automatic classification technology, applied in genomics, sequence analysis, instruments, etc., can solve the problems of 40-60 minutes of time-consuming, time-consuming, and high-level requirements for interpretation technicians, so as to avoid psychological pressure and save money. Operation time, effect of reducing the proportion of undetermined clinical significance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] The embodiment of the present invention takes the BRCA2 gene variation NM_000059.3 (BRCA2):c.4677del (p.Phe1559LeufsTer9) as an example to illustrate the process of using the method of the present invention to interpret it and classify it.
[0075] This variant is an insertion / deletion variant, and the predicted protein consequence is a frameshift, so this variant type is a frameshift variant. The evidence that needs to be judged according to the type of mutation includes at least: PVS1, PM2, PS4, PP5. Evidences PVS1, PM2, PS4, PP5 were searched sequentially in public databases and the literature database. After searching, it was found:
[0076] For PVS1, according to the evidence evaluation rules established in the present invention, it satisfies "when the pathogenic mechanism of a disease is loss of function (LOF), non-functional variants (nonsense mutations, frameshift mutations, classical ± 1 or 2 splicing mutations, Start codon variant, single or multiple exon de...
Embodiment 2
[0085] The embodiment of the present invention takes the BRCA1 gene variation BRCA1:NM_007294.3(BRCA1):c.80+4A>T as an example to illustrate the process of using the method of the present invention to interpret it and classify it. This variant is an intronic variant and cannot be ruled out to have an effect on mRNA splicing. The evidence that needs to be judged according to the type of mutation includes at least: PP3, PM2, PS3. Evidence PP3, PM2, PS3 was searched sequentially in the public database and said literature database. After searching, it is found that: for PP3, according to the evidence evaluation rules established in the present invention, if "when the intron variation satisfies SpliceAI, (anyΔscore≥0.2)", it can be judged that the variation conforms to the evidence of PP3. For PM2, it was retrieved that the mutation was missing in population frequency in gnomAD, which satisfies the judgment rule of the present invention: "PM2_MOD: The occurrence of this mutation w...
Embodiment 3
[0098] The embodiment of the present invention takes the PALB2 gene mutation NM_024675.4 (PALB2): c.1054G>C (p.Glu352Gln) as an example to illustrate the process of using the method of the present invention to interpret it and classify it.
[0099] The variant is a missense variant, and the pathogenic impact of the variant cannot be determined for the time being. The evidence that needs to be judged according to the variant type includes at least: BS1, BP4.
[0100] Evidence BS1, BP4 is searched in turn in the public database and the literature database. After searching, it was found:
[0101] For BS1, according to the evidence judgment rule established by the present invention, it satisfies "In the gnomAD v2.1.1 (non-cancer) database, when the Popmax Filtering AF (95% confidence) corresponding to a certain variation that is qualified for quality control is greater than 0.01% (0.0001 ), then BS1 is satisfied.”
[0102] For BP4, according to the evidence judging rules establi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com