Novel multi-target combined marker for early detection of liver cancer and application thereof
A marker, a technology for liver cancer, applied in measurement devices, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problem of inability to distinguish liver cancer and hepatitis patients, low sensitivity and specificity, and low liver cancer AUC value, etc. problem, to achieve the effect of consistent test results, high test results, and improved effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
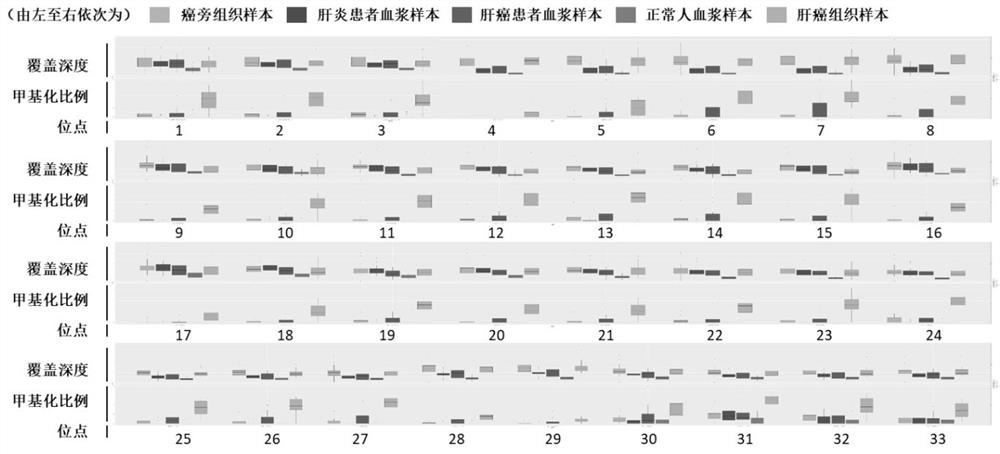
[0089] Example 1 Screening of gene methylation sites
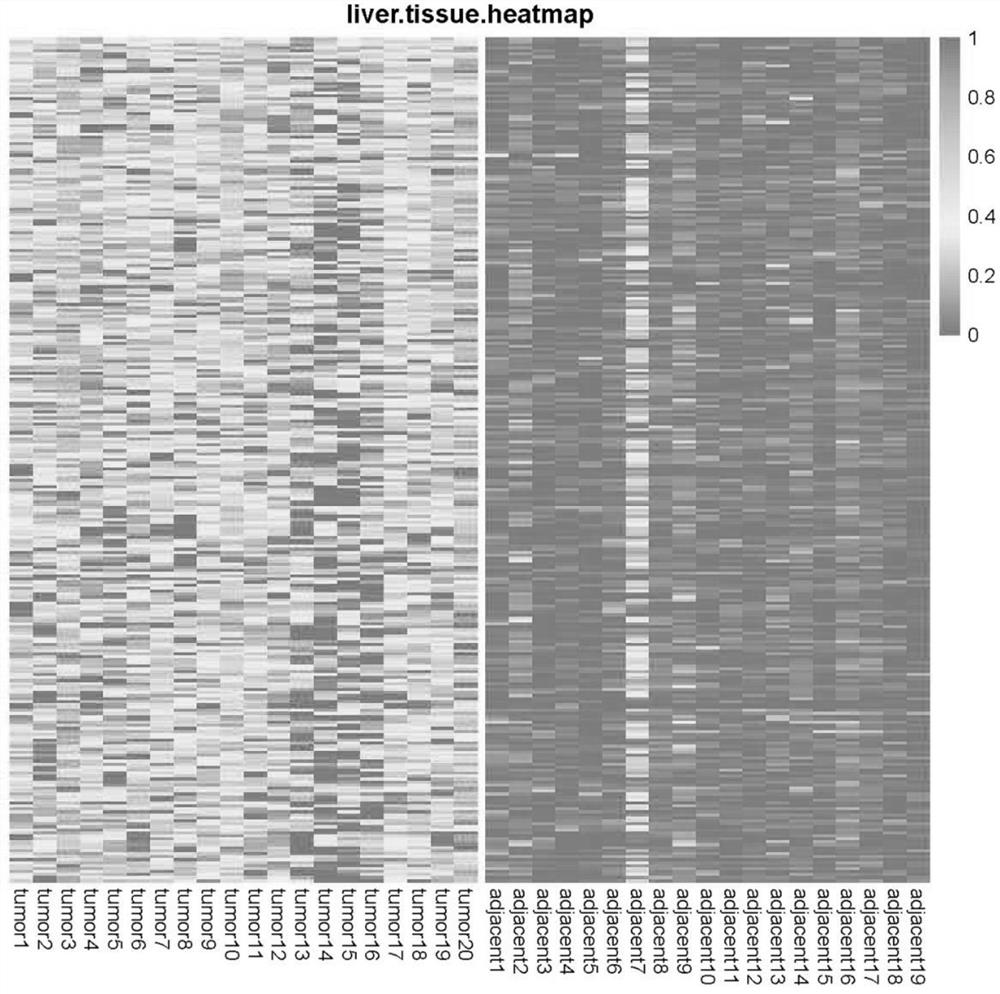
[0090] In this example, high-depth (40×-60×) whole-genome methylation sequencing ( WGBS), selected the differentially methylated regions of 300 genes in the high-depth WGBS data, including both hypermethylated and hypomethylated regions, and efficiently differentiated liver cancer and hepatitis B patients ( figure 1 ). At the same time, a large amount of public data (>1,000 450K microarray data of liver cancer and adjacent cancer cases) was synthesized, and the target region was reduced to 200 genes and 6000 CpG regions.
[0091] In order to avoid the sampling bias of the 200 genes and 6000 CpG regions screened in the previous stage, we further designed a methylation capture panel for these 200 genes, and conducted more than 500 cases of different stages of liver cancer and high-risk groups of liver cancer (hepatitis B and liver cirrhosis). ) cfDNA samples for ultra-high depth (>10,000X) next-generation sequencing to bui...
Embodiment 2
[0093] Example 2 Screening of early liver cancer using 3 methylation sites (or binding protein markers)
[0094] In this example, 3 of the 33 gene methylation sites screened in Example 1 were selected for the detection of early liver cancer. Two methods were used for detection: first, using three methylation sites to detect early-stage liver cancer; second, using a combination of three methylation sites and protein markers to detect early-stage liver cancer.
[0095] 1. Using 3 methylation sites to detect early liver cancer
[0096] The specific method for detecting early-stage liver cancer using three methylation sites includes the following steps:
[0097] In the first step, the serum and plasma of the blood samples were separated, and the magnetic bead method was used to extract the reagents to extract the plasma-free DNA of the biological samples to be tested. Among them, 20 were liver cancer patients and 20 were hepatitis patients.
[0098] In the second step, the methy...
Embodiment 3
[0131] Example 3 Screening of early-stage liver cancer using 6 methylation sites (or binding protein markers)
[0132] In this example, 6 of the 33 gene methylation sites screened in Example 1 were selected for the detection of early liver cancer. Two methods were used for detection: first, using 6 methylation sites to detect early liver cancer; second, using the combination of 6 methylation sites and protein markers to detect early liver cancer.
[0133] 1. Using 6 methylation sites to detect early liver cancer
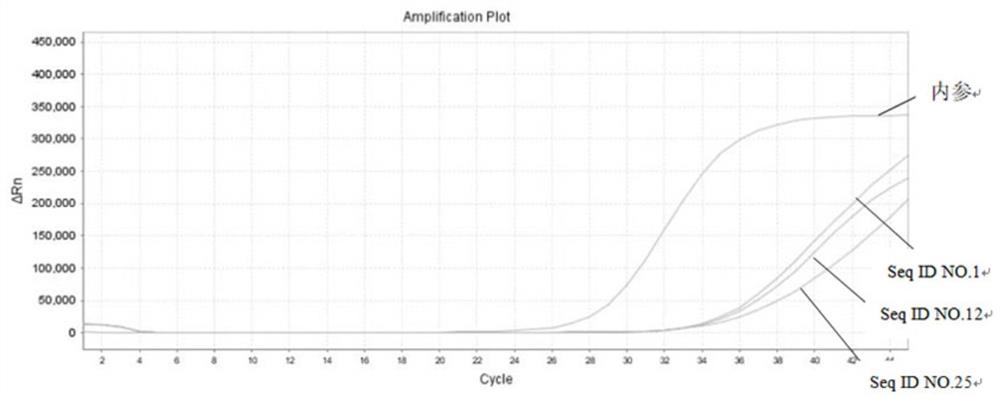
[0134] The present embodiment selects the following groups of combinations comprising 6 methylation sites to detect respectively:
[0135] 4. Seq ID NO.1, Seq ID NO.12, Seq ID NO.13, Seq ID NO.25, Seq ID NO.30, SeqID NO.31
[0136] 5. Seq ID NO.1, Seq ID NO.12, Seq ID NO.23, Seq ID NO.25, Seq ID NO.27, SeqID NO.28
[0137] 6. Seq ID NO.1, Seq ID NO.8, Seq ID NO.12, Seq ID NO.18, Seq ID NO.25, SeqID NO.26
[0138] 7. Seq ID NO.1, Seq ID NO.2, Seq ID NO.3, Seq ID N...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com