Thermolysin variants and detergent compositions therewith
a technology of detergent composition and thermolysin, which is applied in the direction of detergent compounding agent, peptidase, enzymology, etc., can solve the problems of reducing aesthetics, many stains that are difficult to completely remove, and destabilizing the effect of most proteases, so as to improve storage stability and/or catalytic activity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Assays
[0203]The following assays were used in the examples described below. Any deviations from the protocols provided below are indicated in the examples. In these experiments, a spectrophotometer was used to measure the absorbance of the products formed after the completion of the reactions.
A. Bradford Assay for Protein Content Determination in 96-Well Plates
[0204]The Bradford Dye reagent (Quick Start) assay was used to determine the protein concentration in thermolysin samples on a microtiter plate (MTP) scale.
In this assay system, the chemical and reagent solutions used were:[0205]Bradford Quick Start Dye Reagent™ (BIO-RAD Catalogue No. 500-0205)[0206]Dilution Buffer (10 mM NaCl, 0.1 mM CaCl2, 0.005% TWEEN®-80)
The equipment used was a Biomek FX Robot (Beckman Coulter) and a SpectraMAX MTP Reader (type 340; Molecular Devices). MTPs were obtained from Costar (type 9017).
[0207]In the test, 200 μl Bradford Dye Reagent was pipetted into each well, followed by the addition of 15 μl di...
example 2
Thermolysin Production in B. subtilis
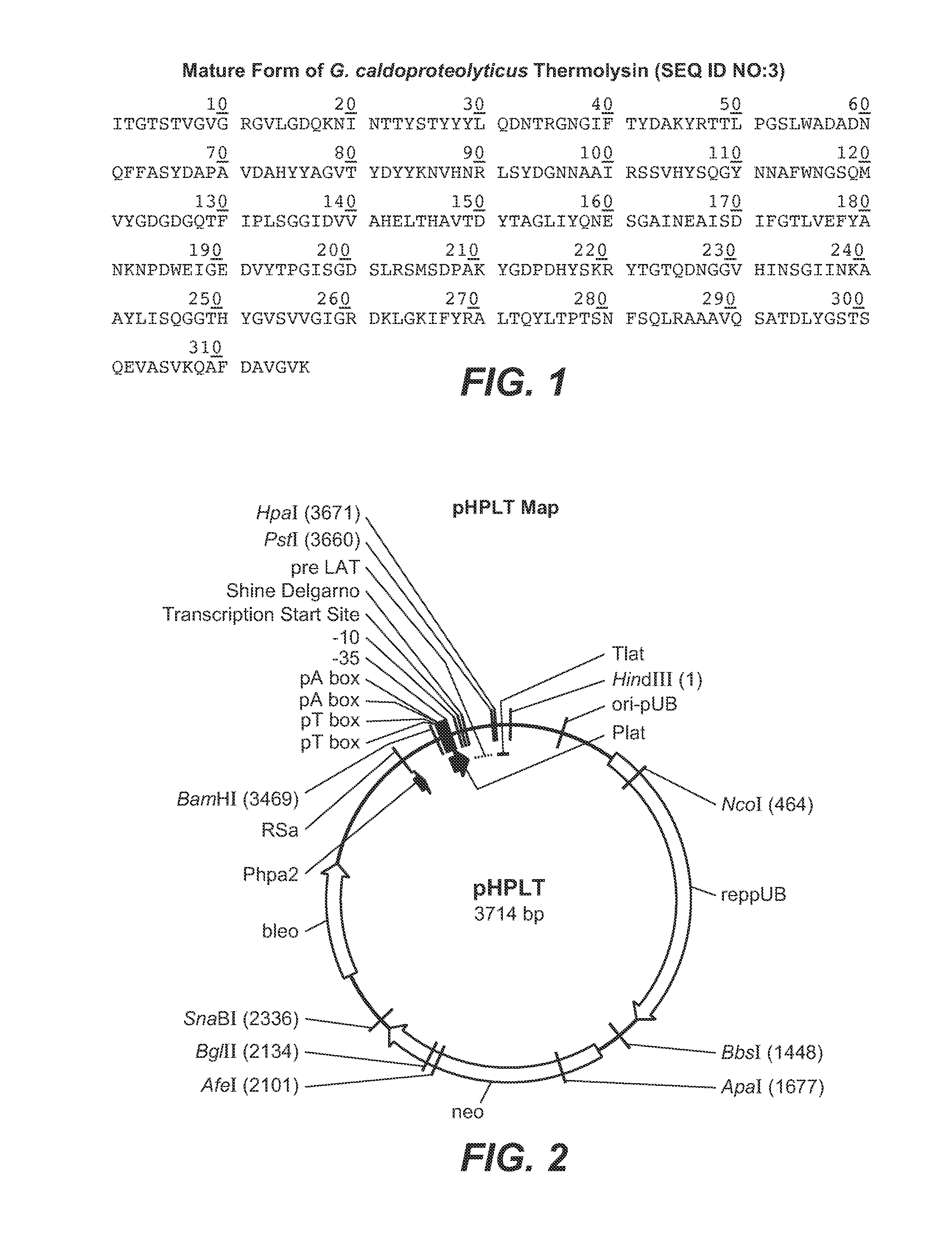
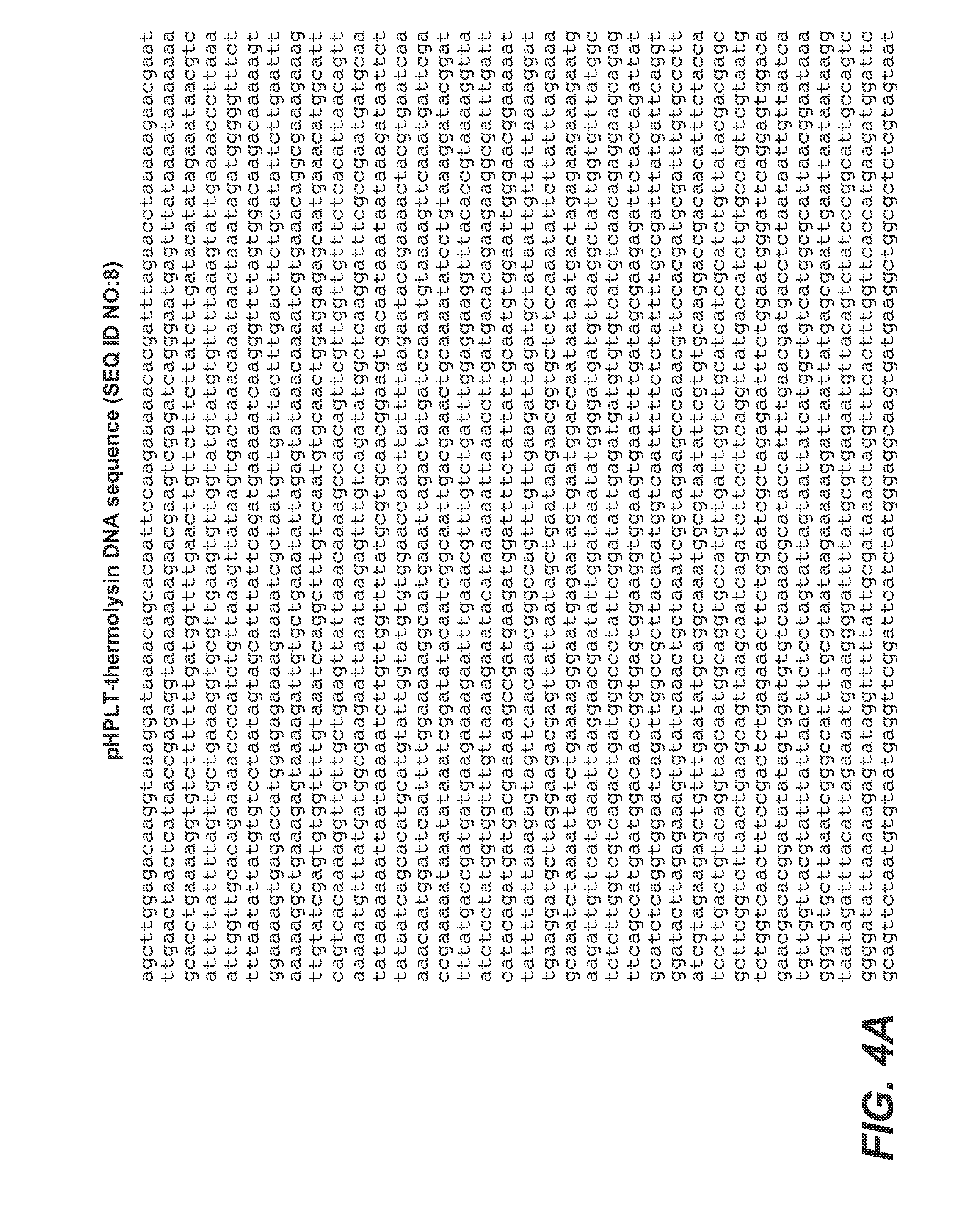
[0238]In this Example, experiments conducted to produce thermolysin in B. subtilis are described. The full-length thermolysin of Geobacillus caldoproteolyticus is greater than 99% identical to the thermolysin precursor of Bacillus thermoproteolyticus Rokko, and to the bacillolysin (NprS) precursor of Bacillus stearothermophilus. As such the terms “thermolysin,”“bacillolysin,”“proteinase-T” and “PrT” are used interchangeably herein to refer to the neutral metalloprotease enzyme of G. caldoproteolyticus. The DNA sequence (thermolysin leader, thermolysin pro and thermolysin mature from Geobacillus caldoproteolyticus) provided below, encodes the thermolysin precursor protein:
(SEQ ID NO: 1)ATGAAAATGAAAATGAAATTAGCATCGTTTGGTCTTGCAGCAGGACTAGCGGCCCAAGTATTTTTACCTTACAATGCGCTGGCTTCAACGGAACACGTTA
[0239]In the above sequence, bold indicates the DNA encoding the mature thermolysin protease, standard font indicates the DNA encoding the leader sequence (thermolys...
example 3
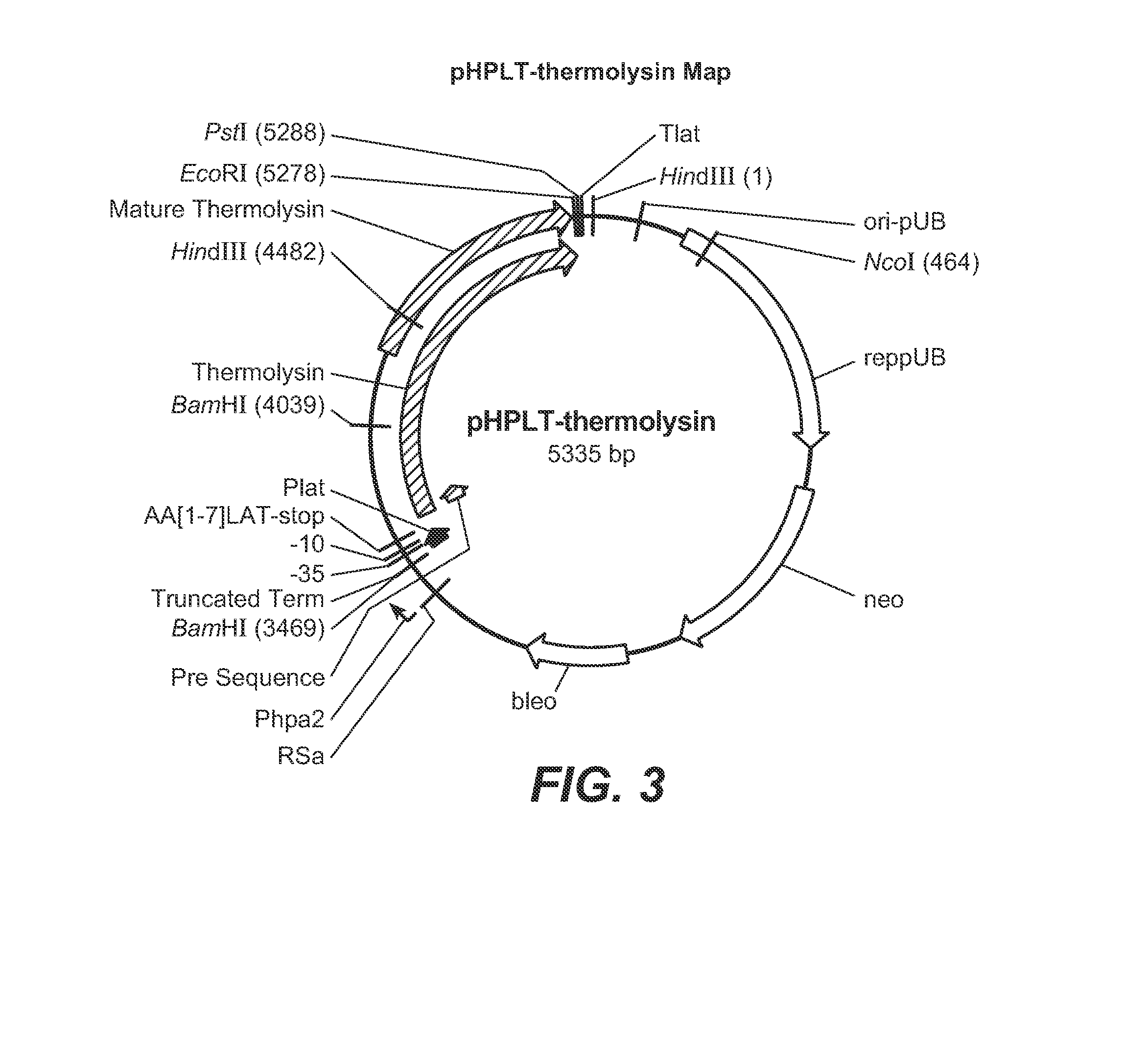
Generation of Thermolysin Site Evaluation Libraries (SELs)
[0242]In this Example, methods used in the construction of thermolysin SELs are described. As previously indicated, the terms “thermolysin,”“bacillolysin,”“proteinase-T” and “PrT” are used interchangeably throughout to refer to the neutral metalloprotease enzyme of G. caldoproteolyticus. The pHPLT-thermolysin vector (FIG. 3) contains the thermolysin expression cassette, which served as a template DNA for the site evaluation libraries. Every thermolysin site evaluation library contains a collection of B. subtilis clones, all expressing a specific thermolysin variant. Each library contains B. subtilis clones, maximally including 20 different variants. For example, thermolysin SEL 27 contains variants in which the DNA triplet coding for tyrosine at position 27 of the mature thermolysin enzyme is replaced by another DNA triplet encoding: Alanine, Aspartic acid, Cysteine, Glutamic acid, Phenylalanine, Glycine, Histidine, Isoleucin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com