Probes for improved melt discrimination and multiplexing in nucleic acid assays
a nucleic acid assay and detection method technology, applied in the field of molecular biology, can solve the problems of limited multiplexing capability of current real-time pcr, difficulty in quantifying the starting template, and unparallel amplification and precision capability, and achieve the effect of increasing the multiplexing capability of real-time nucleic acid amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Hairpin Probe Extension with Unique Melt Signatures
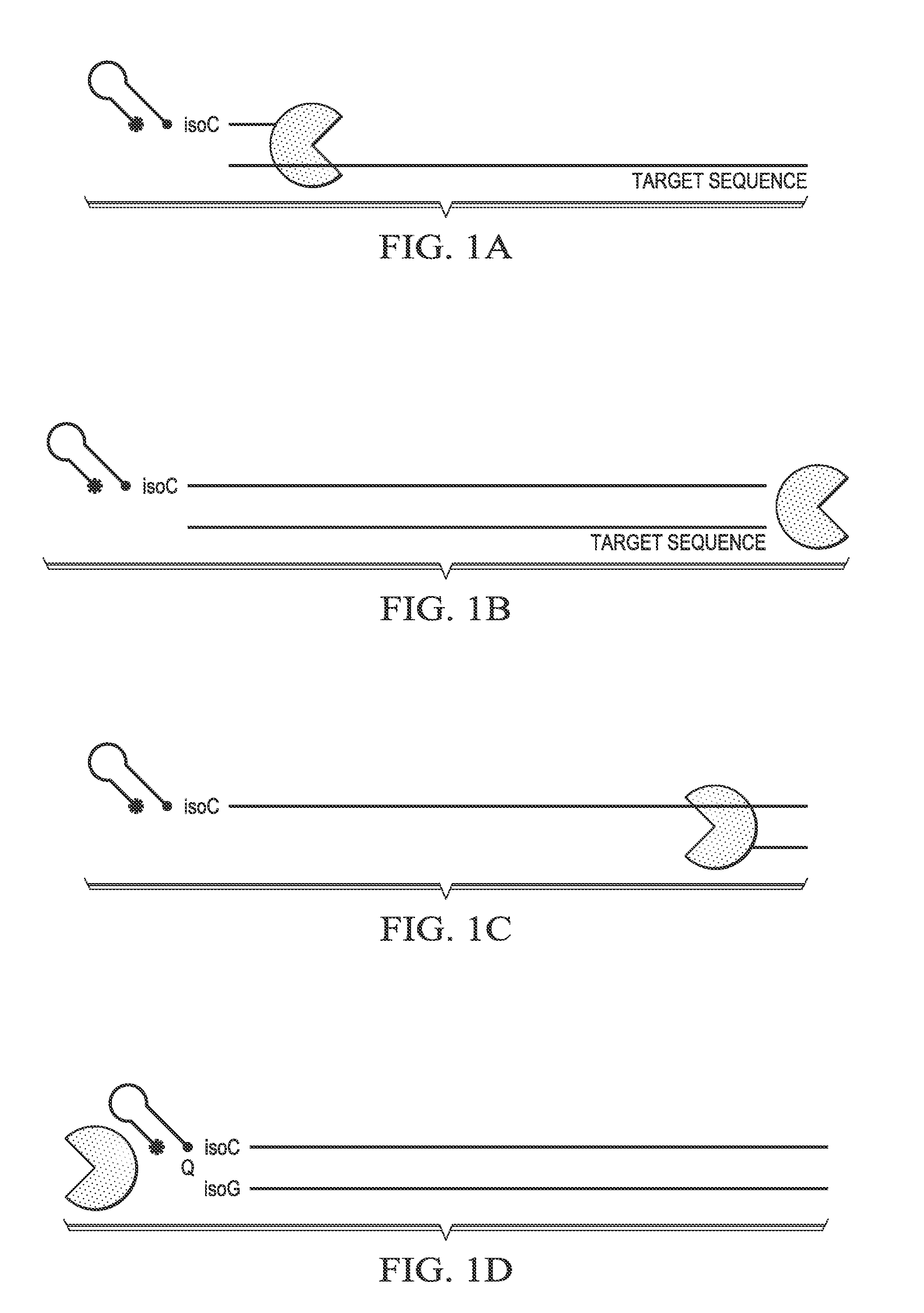
[0147]An extendable probe with a hairpin has been designed to provide unique melt peaks during a melt analysis to allow for greater discrimination of target identity in a melt assay, which can allow for greater multiplexing. This extendable probe can function as a probe when targeted to a sequence within an amplicon for a second level of discrimination, or it can be used as a primer in a primer set (FIGS. 1A-B).
[0148]The extendable probe comprises, in sequence from the 5′ end to the 3′ end, a reporter (star), for example, a fluorophore, a sequence capable of forming a hairpin, a modification designed to block extension (e.g., C3 spacer) during second strand synthesis, an isoG nucleotide, and a target specific sequence. During second strand synthesis using the extended probe as a template, a quencher-labeled isoC nucleotide will be incorporated opposite the isoG nucleotide (FIGS. 1C-D).
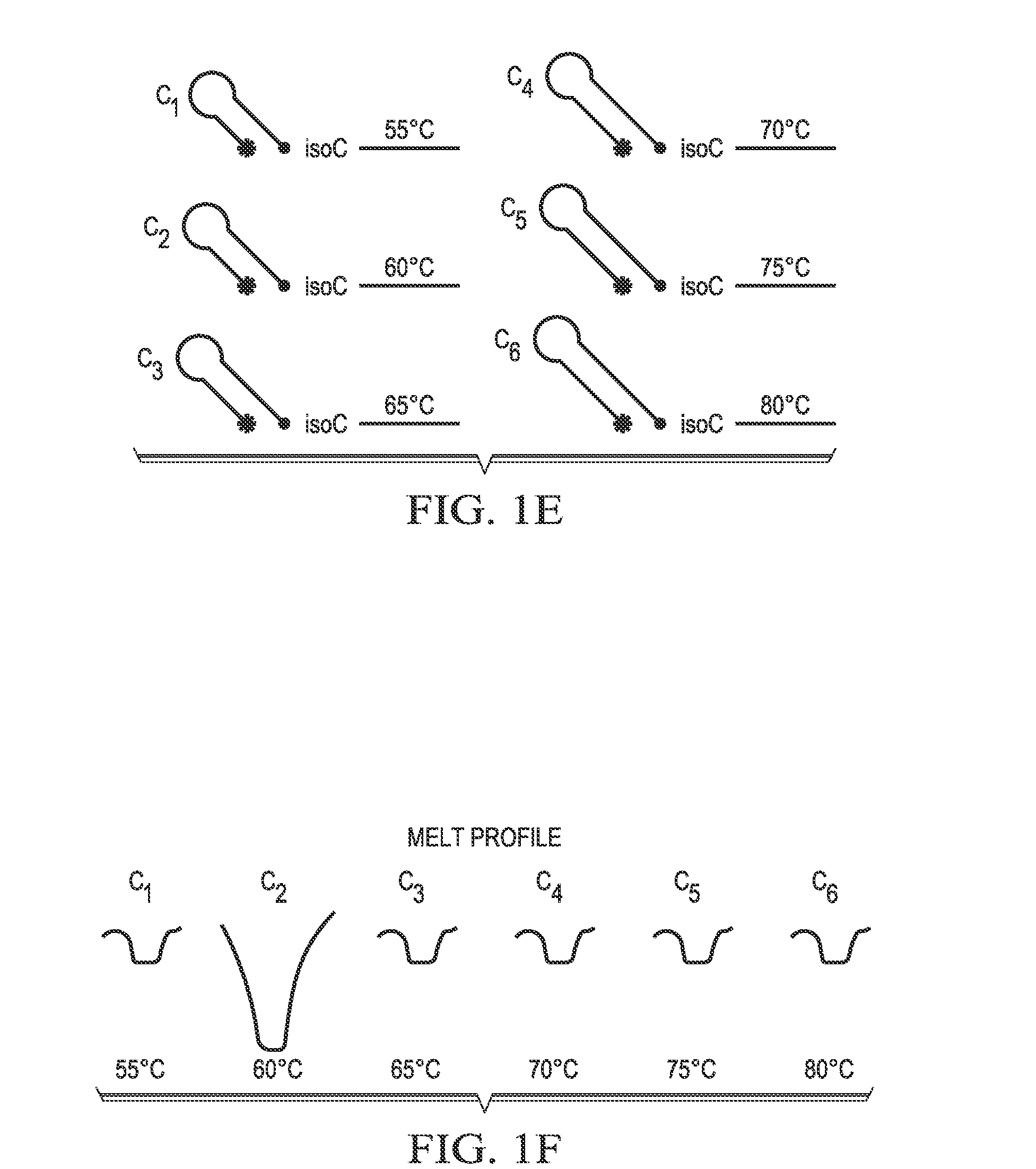
[0149]If hairpin probes are designed such that e...
example 2
Endonuclease Reaction Followed by Extension and Exonuclease Cleavage of Melt-Discrimination Probe-Template Pairs
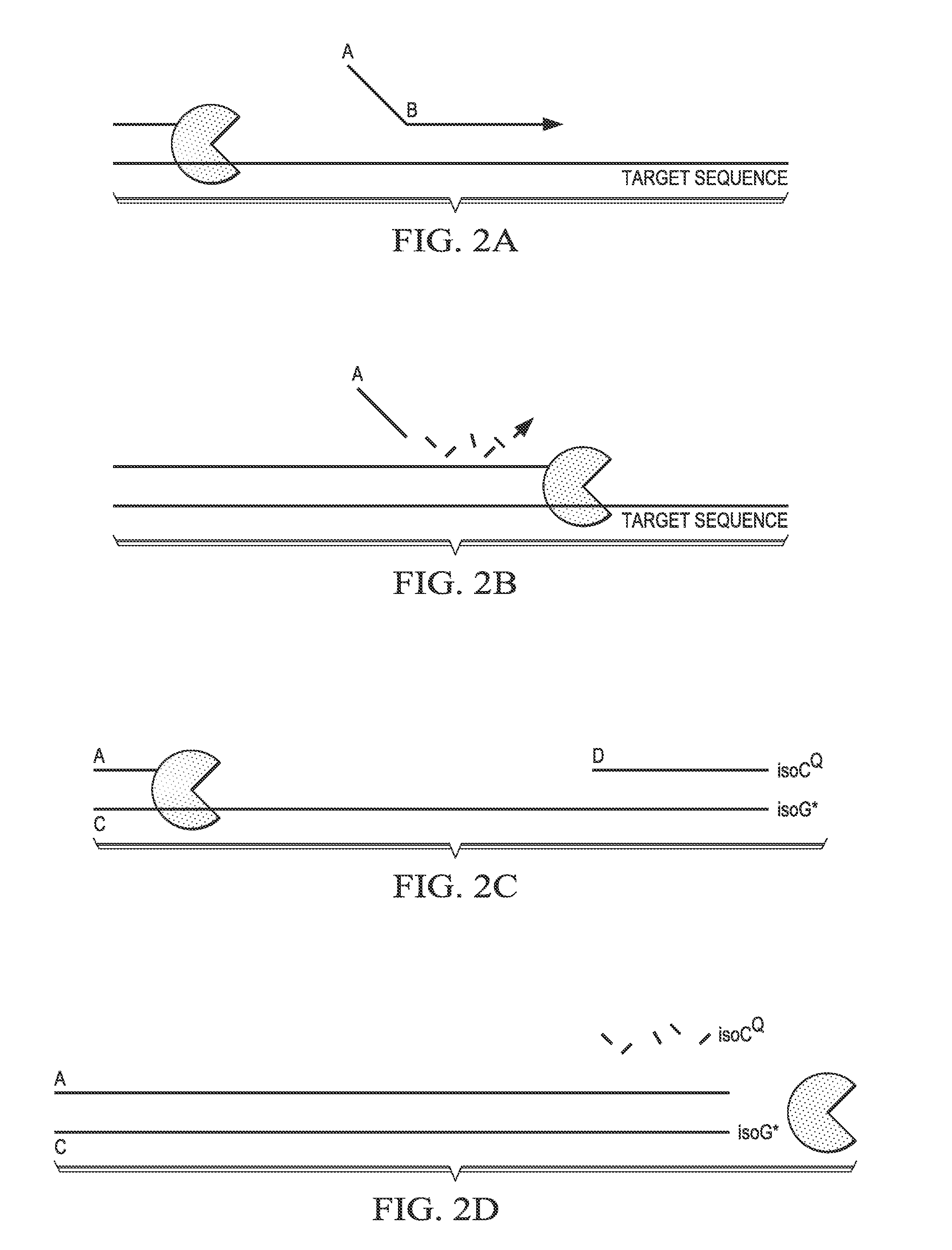
[0152]An extendable probe is designed with a target-specific hybridization sequence at its 3′ end (Sequence B, FIG. 2A) and a melt-discrimination template-specific hybridization sequence at its 5′ end (Sequence A, FIG. 2A). In the presence of the target sequence, polymerase extension from an upstream primer during a PCR reaction cleaves the melt-discrimination template-specific hybridization sequence (FIG. 2B) and degradation of the target-specific hybridization sequence. The cleaved melt-discrimination template-specific hybridization sequence hybridizes to and extends on a melt-discrimination template (Sequence C, FIG. 2C) that has a predesigned melt-discrimination probe (Sequence D, FIG. 2C) hybridized downstream of the binding site for the melt-discrimination template-specific hybridization sequence (FIG. 2C). The 3′ end of the melt-discrimination template comprises a r...
example 3
Endonuclease Reaction Followed by Extension and Exonuclease Cleavage of Melt-Discrimination Probe-Template Pairs that Comprise an Extension Blocker
[0154]An alternative embodiment of the melt-discrimination probe-melt-discrimination template pairs described in Example 2 is presented here. In this embodiment, the melt-discrimination template (Sequence C) comprises a modification designed to block extension (e.g., C3 spacer) with the portion of the template that hybridizes to the melt-discrimination probe (Sequence D) (FIG. 3A). When Sequence A from the hydrolyzed target-specific probe hybridizes to and extends on a melt-discrimination template (Sequence C, FIG. 3A) that has a predesigned melt-discrimination probe (Sequence D, FIG. 3A) hybridized downstream of the binding site for the melt-discrimination template-specific hybridization sequence. The 3′ end of the melt-discrimination template comprises a reporter-labeled isoG nucleotide and the 5′ end of the melt-discrimination probe co...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| Tm | aaaaa | aaaaa |
| Tm | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com