Methods for diagnosing osteoarthritis
a technology of osteoarthritis and prognosis, applied in the field of osteoarthritis prognosis, can solve the problems of incomplete knowledge of the biology of oa, the functional importance of these susceptibility loci has not yet been confirmed,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0138]Comparison of Pitx1 Expression in Articular Chondrocytes of OA Subjects with that of in Articular Chondrocytes Matched Controls
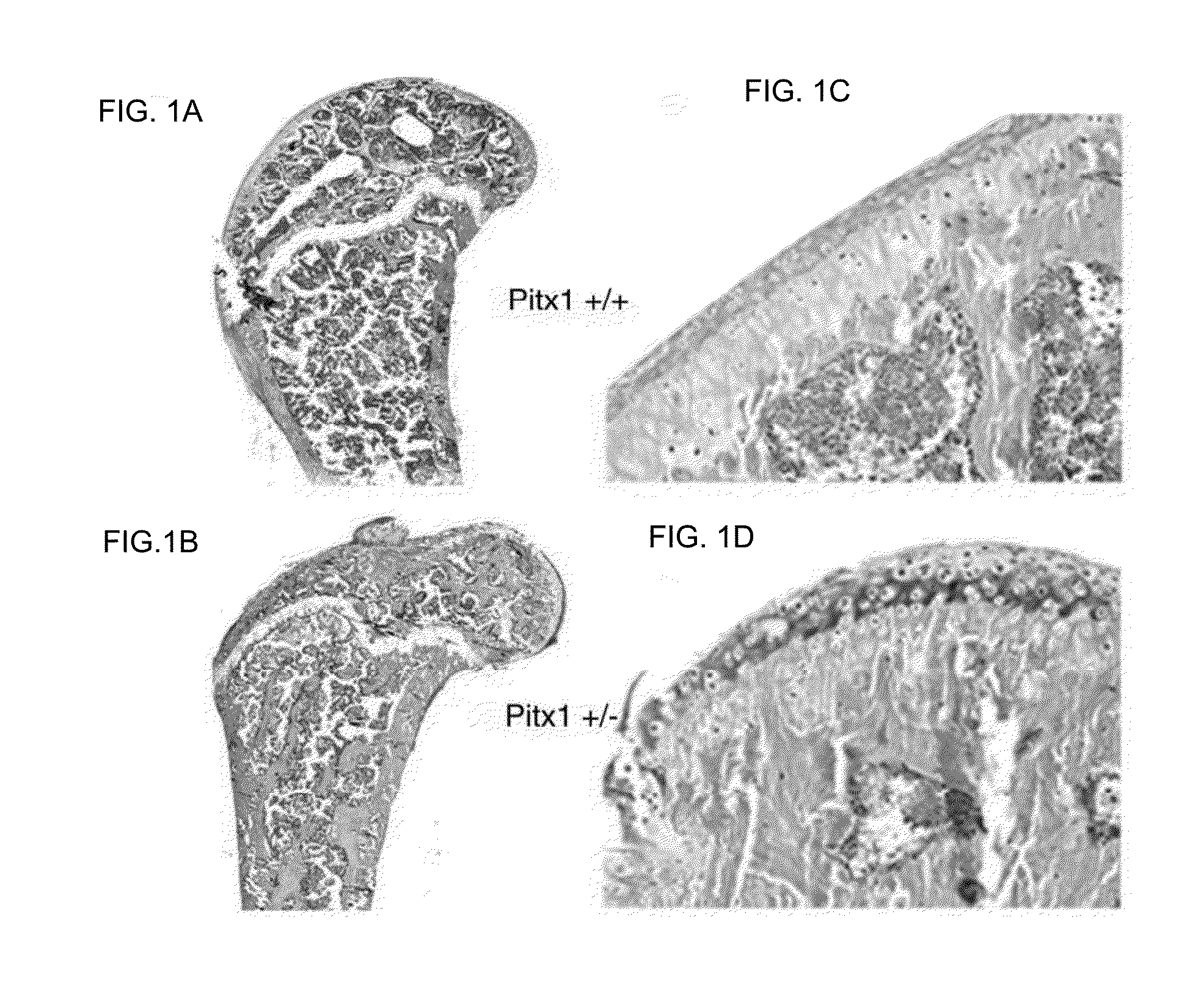
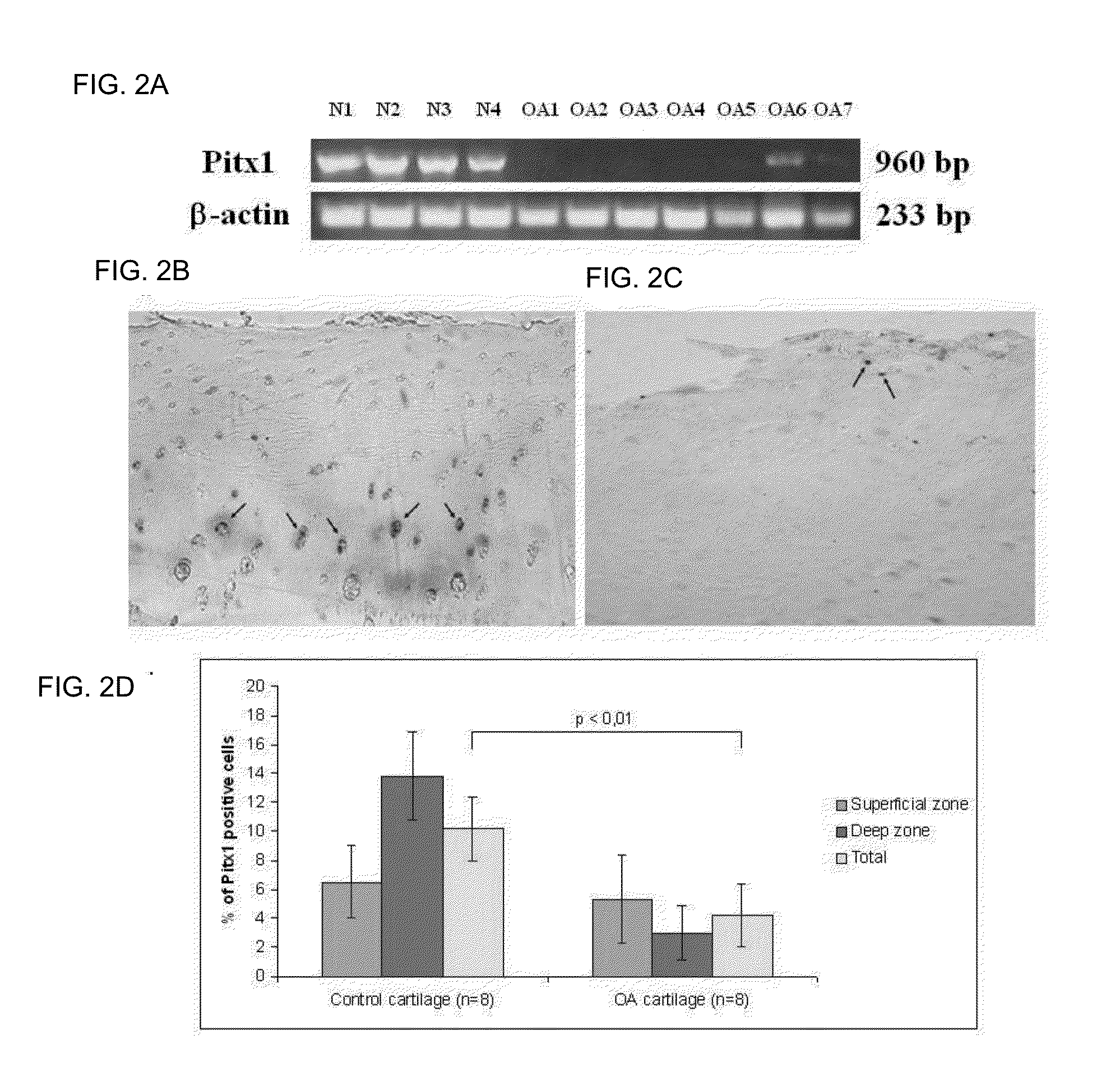
[0139]To determine whether pitx1 plays a role in the genetic control of OA onset, an expression analysis of pitx1 gene using RNA prepared from articular chondrocyte cultures derived from knee cartilage of OA patients (n=7) and age- and gender-matched control subjects (n=4) was performed. Pitx1 expression was detected only in articular chondrocytes derived from matched controls, while in OA articular chondrocytes, Pitx1 expression was abrogated or barely detectable by RT-PCR (FIG. 2A). Analysis of Pitx1 protein levels and distribution in human knee joint sections showed Pitx1 proteins only in control cartilages (n=8), while Pitx1 proteins were hardly detected in OA cartilage sections (n=8) (FIGS. 2B-D).
example 2
Identification of Pitx1 Promoter Mutation
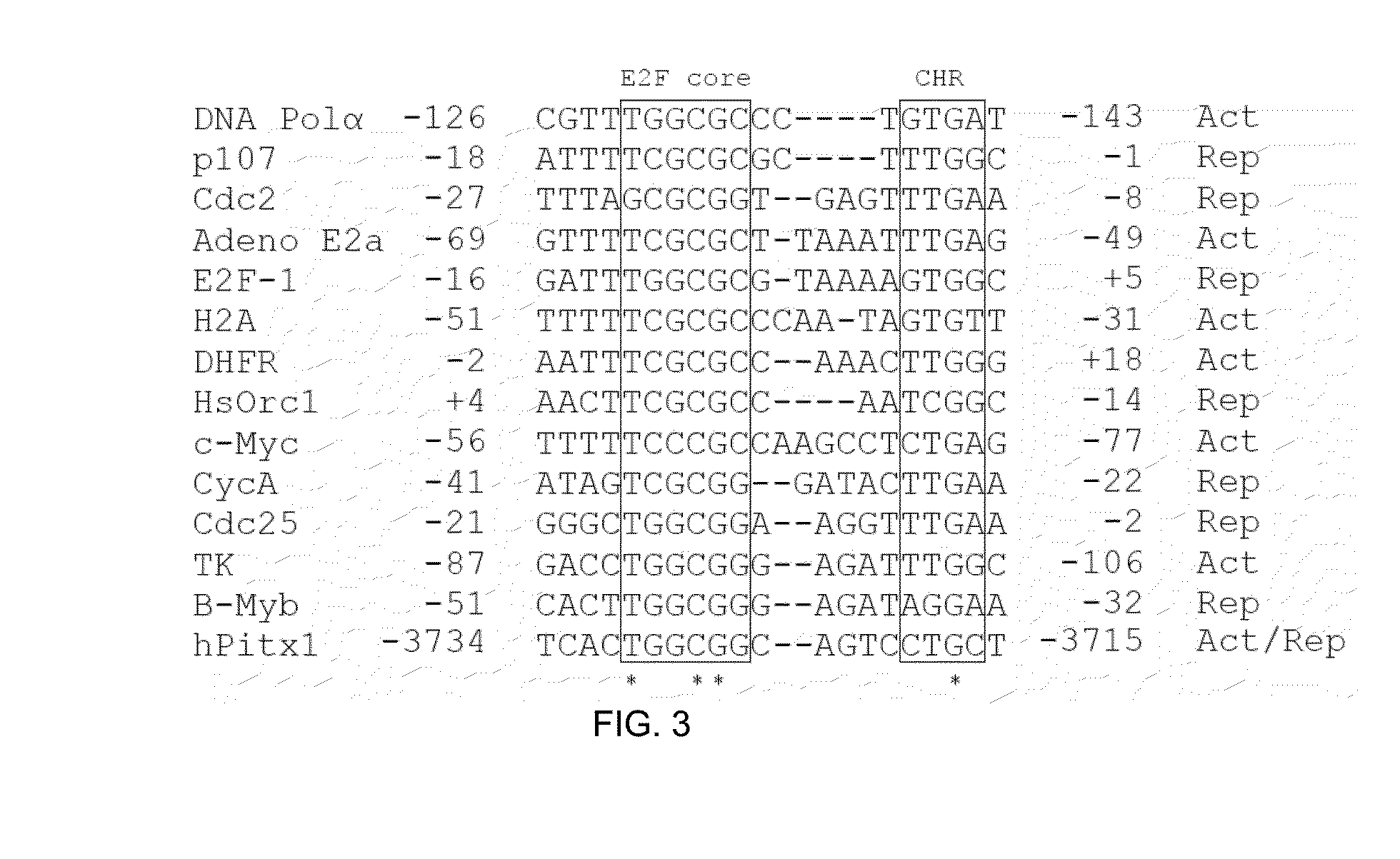
[0140]To examine the mechanisms turning off pitx1 gene expression in OA patients, the 5′ regulatory region of human pitx1 gene was examined for specific mutations leading to a progressive loss of Pitx1 expression during adulthood. Sequencing analysis of genomic DNA obtained from OA, rheumatoid arthritis (RA) and matched control subjects revealed, along a 10 kb promoter region of human pitx1 gene, a single homozygous mutation (−3727 C→T) (position corresponds to distance from transcription point) affecting only OA patients (11 / 43) with a high frequency (25%) while none of the RA patients (0 / 29) and matched control subjects (0 / 11) had the homozygous mutation. The specificity, the positive predictive values and negative predictive values of the mutation were calculated for each group as reported in Table 4 below. A statistically significant association between the mutation and diagnosis was calculated (two-tailed test) by comparing OA versus RA ...
example 3
Determination of Functional Consequences of Mutation in the E2F-Like Site on Complex Binding
[0142]To determine the functional consequences of the homozygous mutation found in OA patients, it was investigated whether E2Fs were able to bind this E2F-like site using nuclear extracts prepared with OA articular chondrocytes as described above. EMSA analysis using both radiolabeled E2F-like sites (wild-type FIG. 4A versus mutant FIG. 4B) showed no supershift of the bound complex with any antibodies against E2Fs, or their dimerization partners DP-1 or DP-2 (E2F2, E2F8 and DP2 data not shown). The Sp1 and Sp3 transcription factors were also analysed since they bind GC-rich regions such as the E2F-like site found in the human Pitx1 promoter. Unfortunately, there was no supershift with either anti-Sp1 or anti-Sp3 antibodies. Addition of BCoR antibodies generated the binding of an additional lower complex bound in presence of either probes although the binding was increased with the mutant one...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thermal melting point | aaaaa | aaaaa |
| thermal melting point | aaaaa | aaaaa |
| thermal melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com