Methods for diagnosing osteoarthritis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Comparison of Pitx1 Expression in Articular Chondrocytes of OA Subjects with that of in Articular Chondrocytes Matched Controls
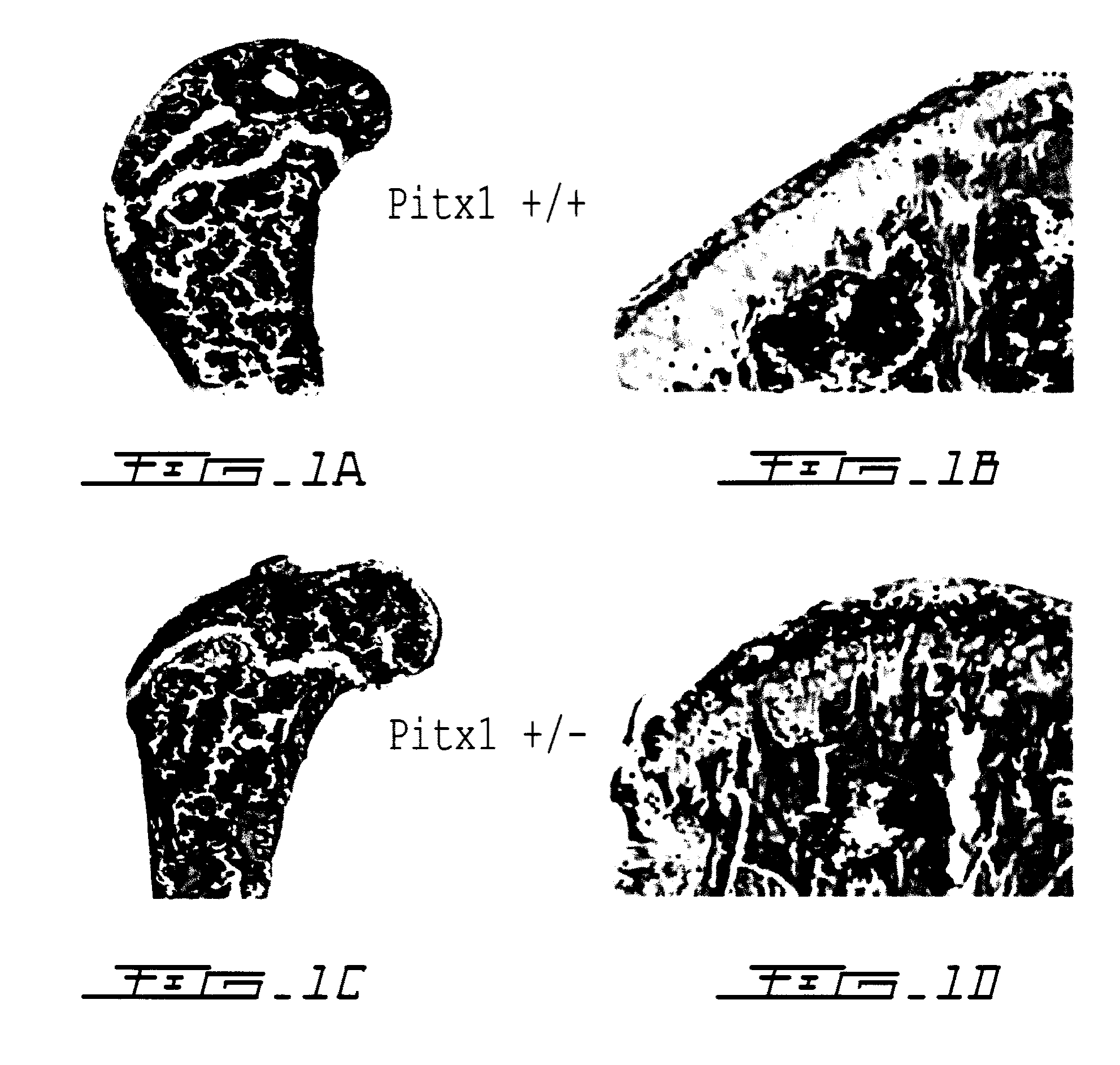
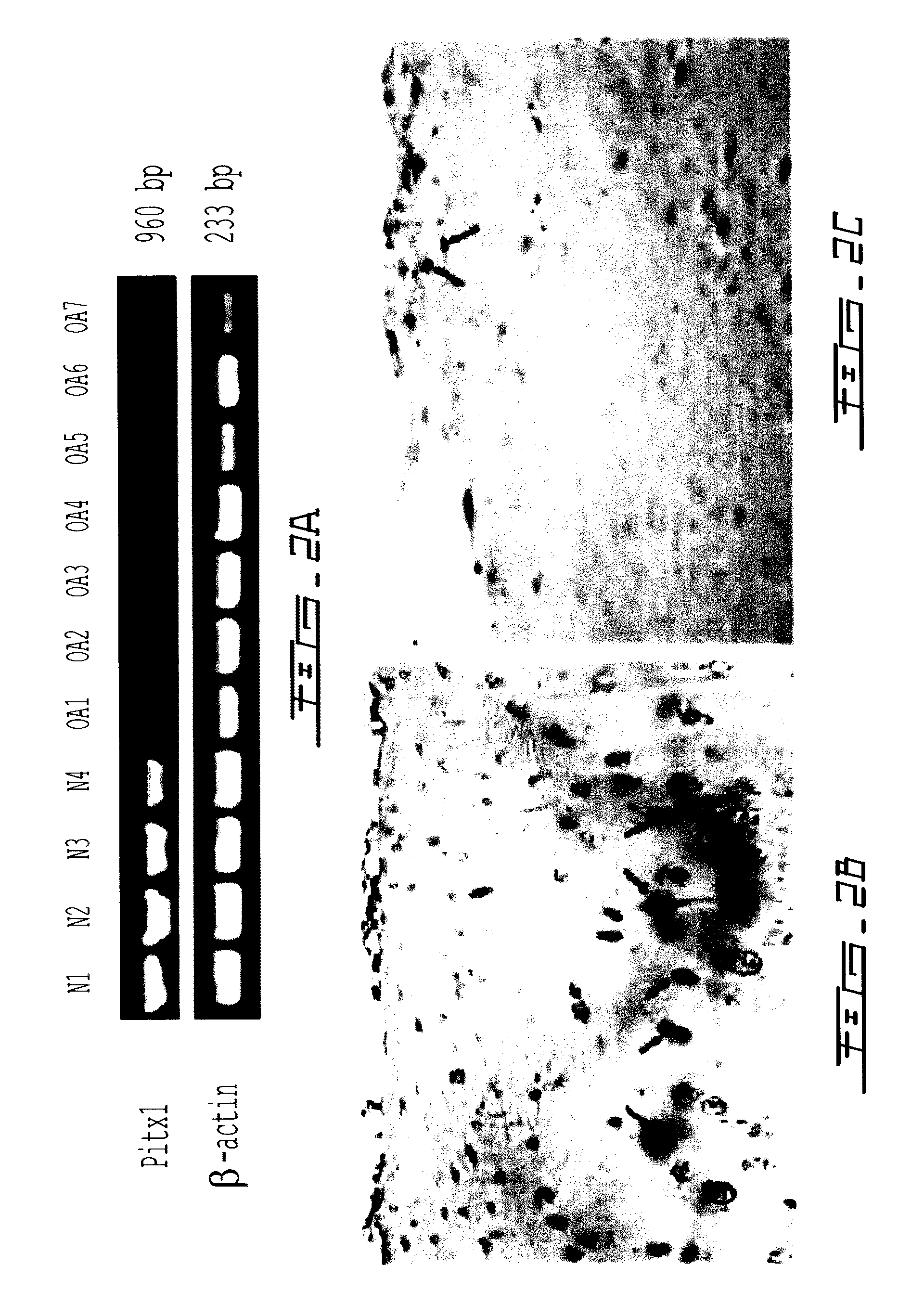
[0138]To determine whether pitx1 plays a role in the genetic control of OA onset, an expression analysis of pitx1 gene using RNA prepared from articular chondrocyte cultures derived from knee cartilage of OA patients (n=7) and age- and gender-matched control subjects (n=4) was performed. Pitx1 expression was detected only in articular chondrocytes derived from matched controls, while in OA articular chondrocytes, Pitx1 expression was abrogated or barely detectable by RT-PCR (FIG. 2a). Analysis of Pitx1 protein levels and distribution in human knee joint sections showed Pitx1 proteins only in control cartilages (n=8), while Pitx1 proteins were hardly detected in OA cartilage sections (n=8) (FIG. 2bcd).
example 2
Identification of Pitx1 Promoter Mutation
[0139]To examine the mechanisms turning off pitx1 gene expression in OA patients, the 5′ regulatory region of human pitx1 gene was examined for specific mutations leading to a progressive loss of Pitx1 expression during adulthood. Sequencing analysis of genomic DNA obtained from OA, rheumatoid arthritis (RA) and matched control subjects revealed, along a 10 kb promoter region of human pitx1 gene, a single homozygous mutation (−3727 C→T) (position corresponds to distance from transcription point) affecting only OA patients (11 / 43) with a high frequency (25%) while none of the RA patients (0 / 29) and matched control subjects (0 / 11) had the homozygous mutation. The specificity, the positive predictive values and negative predictive values of the mutation were calculated for each group as reported in Table 4 below. A statistically significant association between the mutation and diagnosis was calculated (two-tailed test) by comparing OA versus RA ...
example 3
Determination of Functional Consequences of Mutation in the E2F-Like Site on Complex Binding
[0141]To determine the functional consequences of the homozygous mutation found in OA patients, it was investigated whether E2Fs were able to bind this E2F-like site using nuclear extracts prepared with OA articular chondrocytes as described above. EMSA analysis using both radiolabeled E2F-like sites (wild-type FIG. 4a versus mutant FIG. 4b) showed no supershift of the bound complex with any antibodies against E2Fs, or their dimerization partners DP-1 or DP-2 (E2F2, E2F8 and DP2 data not shown). The Sp1 and Sp3 transcription factors were also analysed since they bind GC-rich regions such as the E2F-like site found in the human Pitx1 promoter. Unfortunately, there was no supershift with either anti-Sp1 or anti-Sp3 antibodies. Addition of BCoR antibodies generated the binding of an additional lower complex bound in presence of either probes although the binding was increased with the mutant one...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com