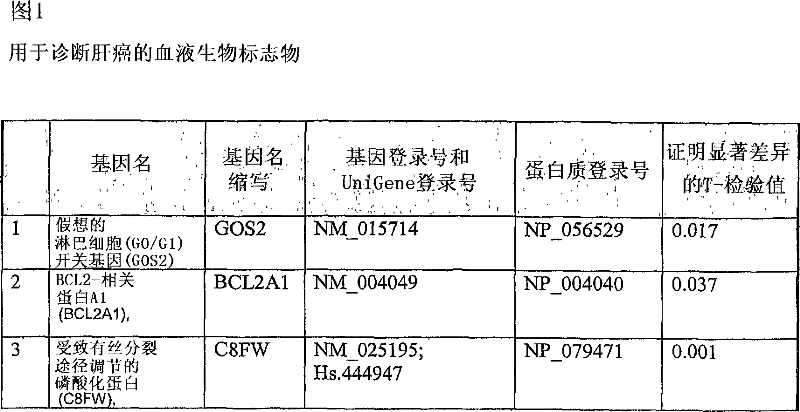
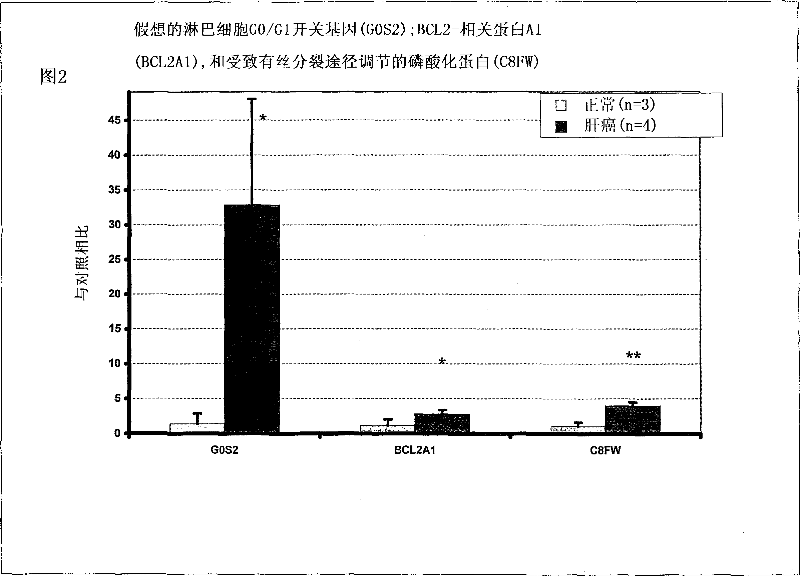
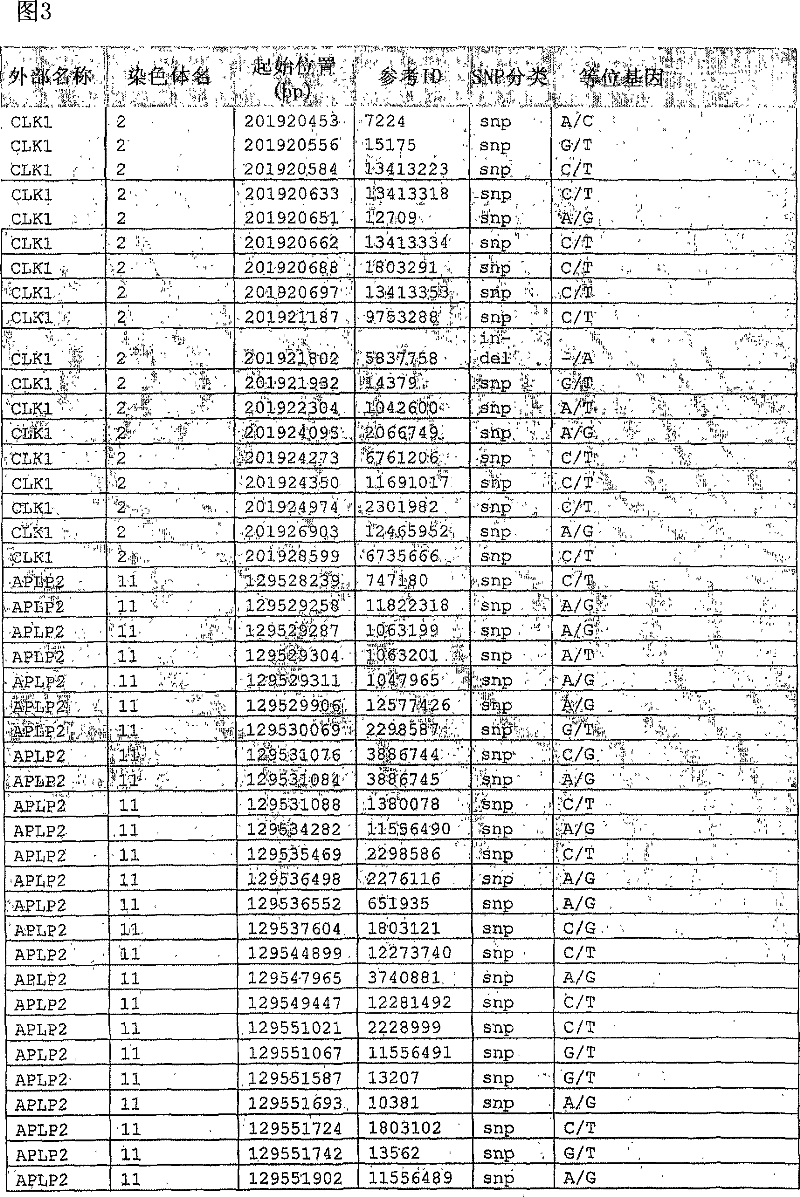
Liver cancer biomarkers
A technology for biomarkers and products, which can be used in sugar derivatives, biochemical equipment and methods, and the determination/inspection of microorganisms, and can solve problems such as easy bleeding
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0606] RNA isolation from lysed blood
[0607] 10 ml of whole blood was collected in a Vacutainer and centrifuged at 2,000 rpm for 5 minutes at 4°C to remove the plasma layer. Add lysis buffer at a ratio of 3 parts lysis buffer to 1 part blood (lysis buffer (1 L): 0.6 g EDTA; 1.0 g KHCO 2 , 8.2g NH 4 Cl, adjusted to pH 7.4 with NaOH). Mixed samples were placed on ice for 5-10 minutes until clear. The lysed samples were centrifuged at 1000 rpm for 10 min at 4°C and the supernatant was discarded by aspiration. The pellet was resuspended in 5 ml of lysis buffer and the lysed sample was centrifuged again at 1000 rpm for 10 min at 4°C. The cells were homogenized with TRIzol_(GIBCO / BRL) at a ratio of approximately 6 ml TRIzol_ per 10 ml of original blood sample and shaken well. The samples were left to stand at room temperature for 5 minutes. RNA was extracted with 1.2ml chloroform / 1ml TRIzol®. Centrifuge the samples at 12,000 x g for 5 min at 4 °C and collect the upper layer...
Embodiment 2
[0609] RNA isolation from whole blood
[0610] 100 μl of whole blood was collected in a microcentrifuge tube, centrifuged at 2,000 rpm (800 g) at 4° C. for 5 minutes, and the supernatant was removed. The cells were homogenized with TRIzol (GIBCO / BRL) at a ratio of approximately 6 μl TRIzol per 10 μl of original blood sample and shaken thoroughly. The samples were left to stand at room temperature for 5 minutes. RNA was extracted with 12 μl chloroform / 10 μl TRIzol. Centrifuge the samples at 12,000 x g for 5 min at 4 °C and collect the upper layer. Add isopropanol to the upper layer at a ratio of 5 μl / 10 μl TRIzol®. Samples were left at -20°C overnight or at -20°C for 1 hour. Precipitate RNA according to known methods, air dry the RNA pellet, and resuspend the pellet in DEPC-treated ddH 2 O middle. RNA samples can also be stored in 75% ethanol, under which conditions the samples are stable at room temperature for shipping.
[0611] RNA isolation from centrifuged lysed blo...
Embodiment 3
[0616] Target nucleic acid preparation and hybridization
[0617] Preparation of fluorescent DNA probes from mRNA
[0618] Fluorescently labeled target nucleic acid samples of RNA are prepared for analysis with the arrays of the invention.
[0619] 1 μg of Oligo-dT primer was annealed to 10 μg of total RNA in a total volume of 10 μl obtained from the blood of patients diagnosed or suspected of having liver cancer by heating at 70 °C for 10 min and cooling on ice. separate. Incubate samples at 42°C for 40 minutes to reverse transcribe mRNA in a 25 μl volume containing final concentrations of 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl 2 , 25 mM DTT, 25 mM unlabeled dNTPs, 400 units of Superscript II (200 U / μl, BRL) and 15 mM Cy3 or Cy5 (Amersham). The reaction was stopped by adding 2.5 μl of 55500 mM EDTA and 5 μl of 1 M NaOH and incubating at 65° C. for 10 minutes. The reaction mixture was neutralized by adding 12.5 μl of 1M TrisHCl (pH 7.6).
[0620] Labeled target nuc...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com