Antibody library screening method
A library and antibody technology, applied in chemical libraries, library screening, anti-tumor drugs, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0197] Example 1 - Testing the BRASIL method
[0198] After incubation with the phage library for 1-2 hours, the two breast cancer cell lines and PBL were centrifuged through the organic phase, and after centrifugation of the organic phase, they were separated in the aqueous layer (upper phase) and the organic layer. (agglomerates) for detection. As a negative control, 10 11 Phage were centrifuged to assess the amount of free phage entering the organic phase.
[0199] Recoveries after panning of human tumor cell lines (PM-1 and MT-1 ) and PBLs were examined. Indicates that PBL must be freshly isolated. Thawed PBL from frozen aliquots tends to become sticky during the elutriation step. After centrifugation, 75% (PM-1), 97% (MT-1) and 71% (PBL) were determined in the cell pellet in the organic phase. Its percentage is too high to be biologically significant: one would expect that only a small percentage of the entire natural antibody library will bind to cell surface protei...
Embodiment 2
[0201] Example 2 - Comparison of Four Methods for Isolating Episomal Phage from Cell-Associated Phage
[0202] 1) Single-step organic phase separation based on the BRASIL method (Giordano et al., 2001):
[0203] After elutriation, 2×150 μl of tumor cell suspension was directly dissolved in 2×300 μl of organic phase solution (dibutyl phthalate:cyclohexane 9:1 [v:v]; d=1.03 gml -1 ) on top of the layer. After centrifugation at 10,000 g for 10 min, the tube was snap frozen in liquid nitrogen at room temperature (RT), then the bottom of the tube was cut off and the cell pellet was transferred to another (blocked) tube.
[0204] 2) As method 1, but with one or two washing steps with 1 ml PBS / 0.4 BSA before resuspending in 300 μl PBS / 0.4% BSA and centrifuging the organic phase.
[0205] 3) Classic method:
[0206] After elutriation, the tumor cells were pelleted by centrifugation (500 g, 5 min, 4° C.), and the pellet was washed 5 times with 1 ml PBS / 0.4% BSA.
[0207] 4) Magneti...
Embodiment 3
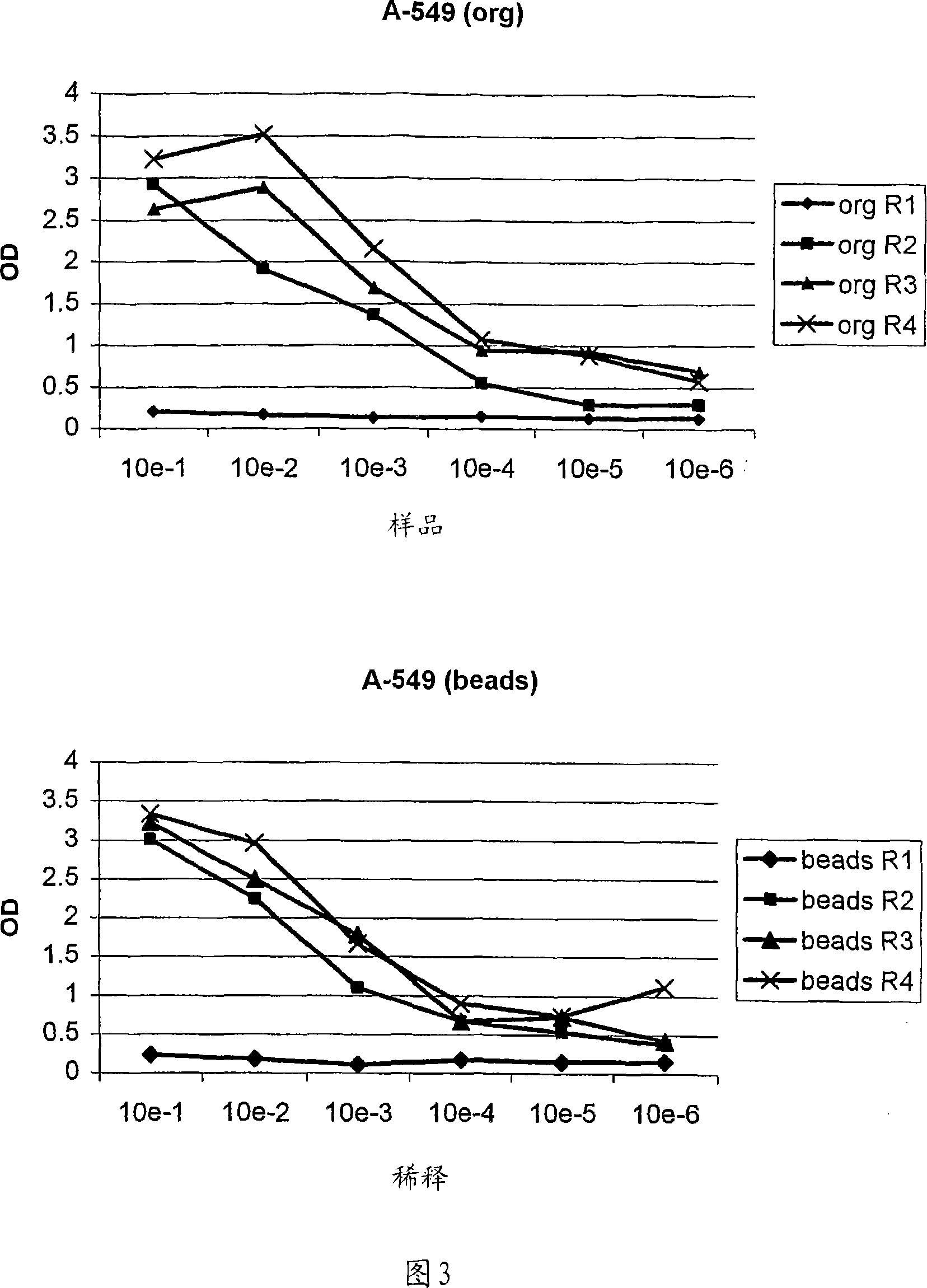
[0216] Example 3 - Successful elutriation protocol with organic phase separation after 1 or 2 washes
[0217] Both elutriation protocols successfully employed a combination of 1 or 2 washes and organic phase separation.
[0218] 1) Breast Cancer Program
[0219] A breast cancer cell line (PM-1) was used to pan the phage scFv display library. Negative pre-panning was performed with fresh human PBL as described in the Materials and Methods section. In these experiments, the effect of washing before centrifugation of the organic phase and its comparison with classical methods (see above) was investigated.
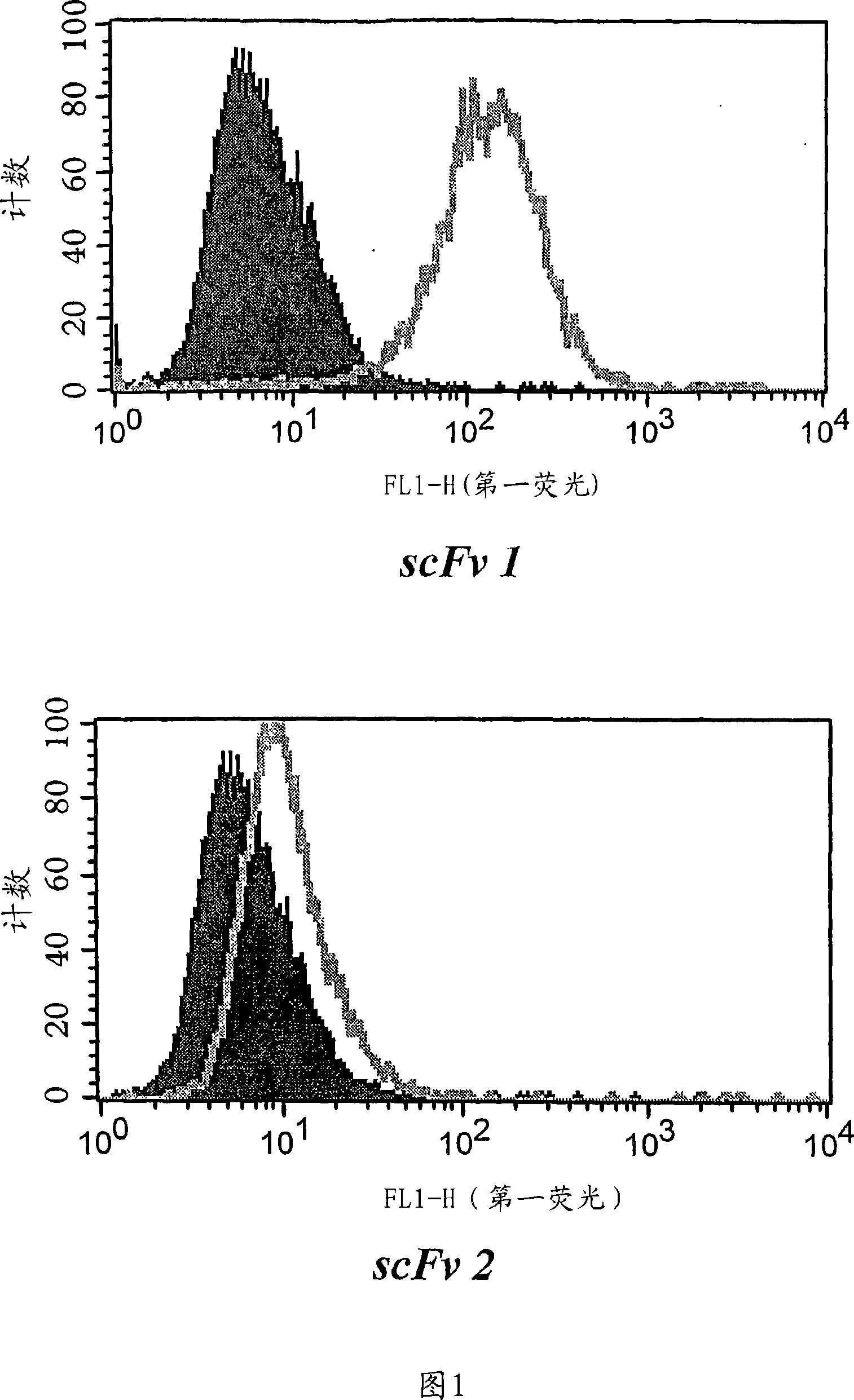
[0220] After one wash and centrifugation of the organic phase, individual phages and scFvs from a third round of panning were tested for specificity in cellular ELISA and FACS measurements relative to PBL and the tumor cell lines. Three unique, tumor cell line-specific clones were found (clones 1, 2 and 3). The expression of single scFv was tested with 8200 cell detection ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com