Recombined kluyveromyces, construction method and applications thereof
A yeast and recombinant bacteria technology, applied in recombinant DNA technology, chemical instruments and methods, microorganism-based methods, etc., can solve the problems of complex, uneven protein purification, and reduced protein specific activity, and avoid high immunogenicity. , The effect of reducing the number of monosaccharides and not easy to reverse mutation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
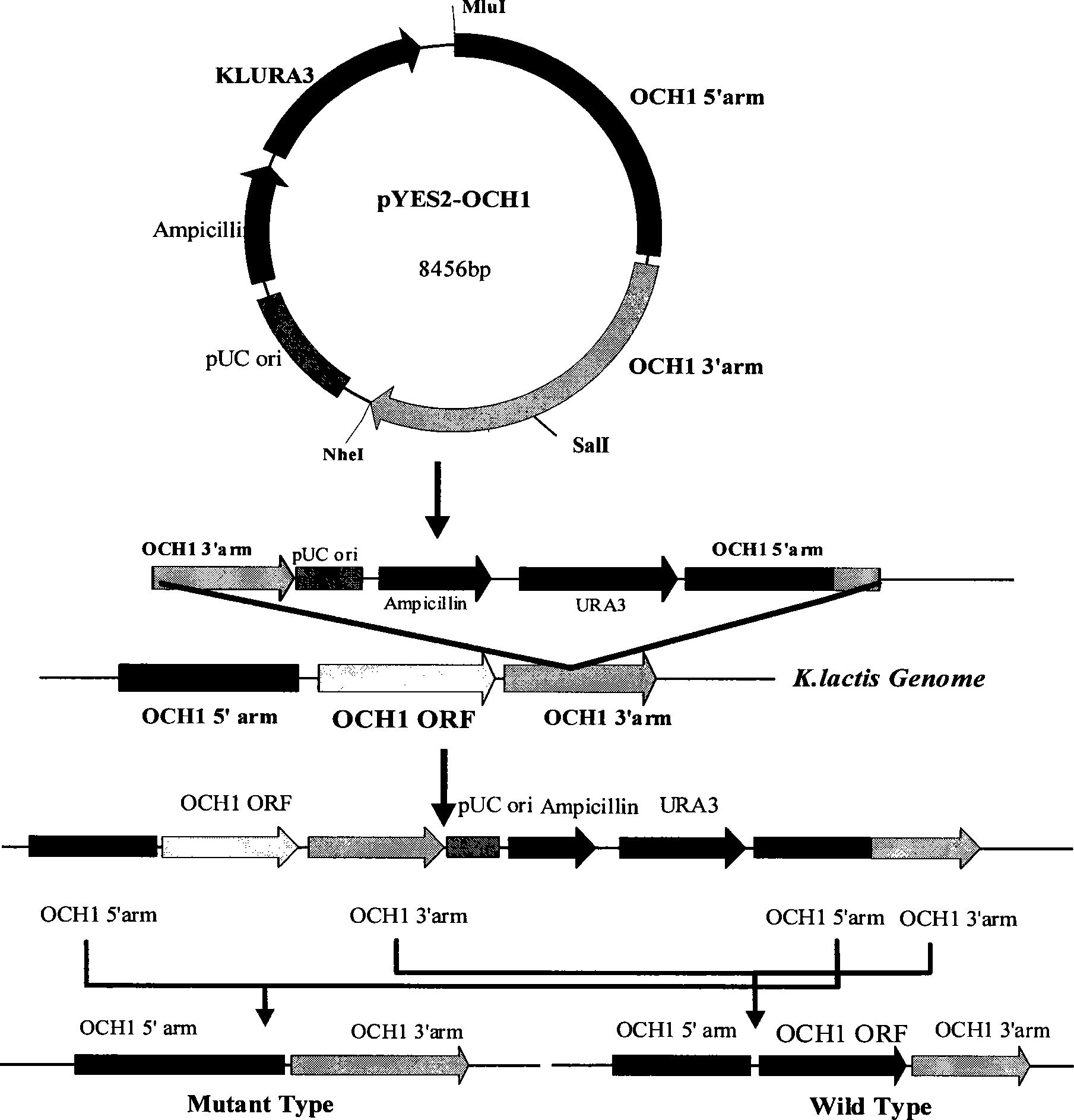
[0064] Example 1, Construction of recombinant Kluyveromyces lactis CGMCC No.2400 inactivated by α-1,6-mannosyltransferase
[0065] 1. Construction of recombinant vector for inactivating α-1,6-mannosyltransferase gene
[0066] Pyrobest enzyme, LA Taq enzyme, dNTPs, restriction endonuclease, T4 ligase, etc. used in the experiment were purchased from Dalian Bao Biological Engineering Co., Ltd., pfu enzyme, kit, DH5α competent cells, primer synthesis, sequencing, etc. were purchased from Provided by Shanghai Sangon Bioengineering Technology Service Co., Ltd.
[0067] Kluyveromyces lactis ATCC8585 (American Type Culture Collection, American Type Culture Collection, Manassas, Va. 20108 USA), using the genomic DNA as a template to amplify the homology arms on both sides of the α-1,6-mannosyltransferase (OCH1) gene (sequence 1 in the sequence listing), and the homology arms on both sides of OCH1 They are 2kb respectively, and the coding gene of about 1.1kb is missing in the middle. ...
Embodiment 2
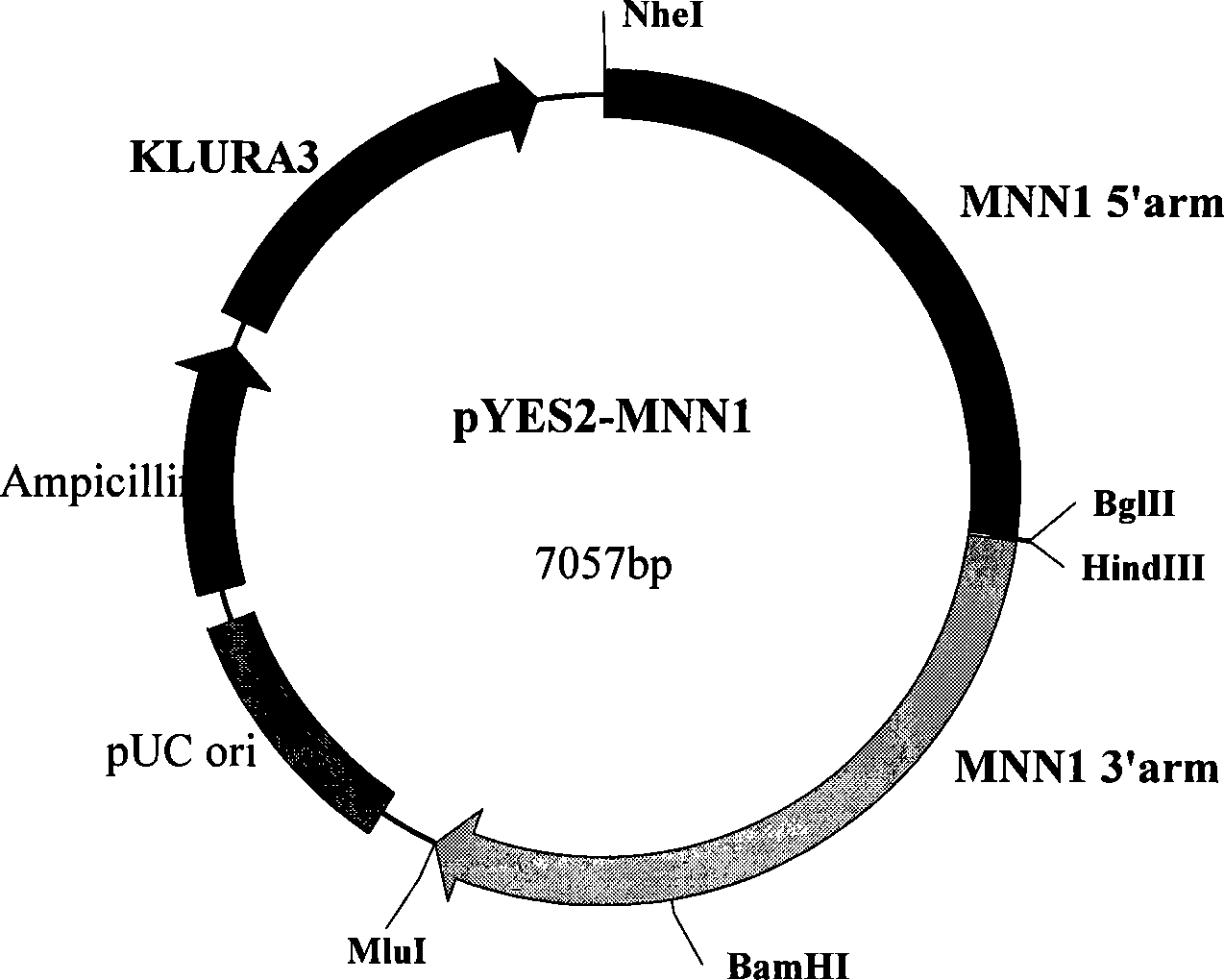
[0079] Example 2, Construction of recombinant Kluyveromyces lactis inactivated by α-1,3-mannosyltransferase
[0080] 1. Construction of recombinant vector for inactivating α-1,3-mannosyltransferase gene
[0081] The genomic DNA of Kluyveromyces lactis ATCC8585 was extracted by the glass bead preparation method (A. Adams et al., "Experimental Guidelines for Yeast Genetics Methods", Science Press, 2000), and the genomic DNA was used as a template to amplify α-1,3 -The homology arms on both sides of the mannosyltransferase gene (sequence 2 in the sequence listing), the homology arms on both sides of the α-1,3-mannosyltransferase gene (MNN1) are 1.4kb and 1.5kb respectively, and the middle is deleted The 2.4kb coding gene, the primers used to amplify the homology arms of the upstream flank region of MNN1 are KLMNN01 and KLMNN02, and the primer sequences are: 5′-TAA GCTAGC TGACCAATGACGCATCCGAACT-3' (the underlined part is the NheI recognition site) and 5'-ATT AAGCTT ATGGCACGCTG...
Embodiment 3
[0092] Example 3: Construction of recombinant Kluyveromyces lactis CGMCC No.2401 in which both α-1,6-mannosyltransferase gene and α-1,3-mannosyltransferase gene were inactivated
[0093] Using recombinant Kluyveromyces lactis CGMCC No.2400 as a host, according to the method of Example 2, inactivating the α-1,3-mannosyltransferase gene in the genome of recombinant Kluyveromyces lactis CGMCC No.2400, at 8 Four mutant strains with inactivated α-1,6-mannosyltransferase gene and α-1,3-mannosyltransferase gene were obtained from the clones transfected with pYES2-mnn1 linear fragment. One strain was named KLGE03. The bacterial strain was deposited in the General Microorganism Center of China Committee for Culture Collection of Microorganisms (abbreviated as CGMCC, address: Datun Road, Chaoyang District, Beijing) on March 17, 2008, and the preservation number is CGMCC No.2401.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com