Biological information recognition method based on dynamic sample selection integration
A biological information identification and biological information technology, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve the problems of not being able to adjust the relationship between the recognition rate of small samples according to needs, the lack of training data, and the impact, etc., to achieve The effect of effectively dealing with biometric information identification problems and reducing time overhead
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
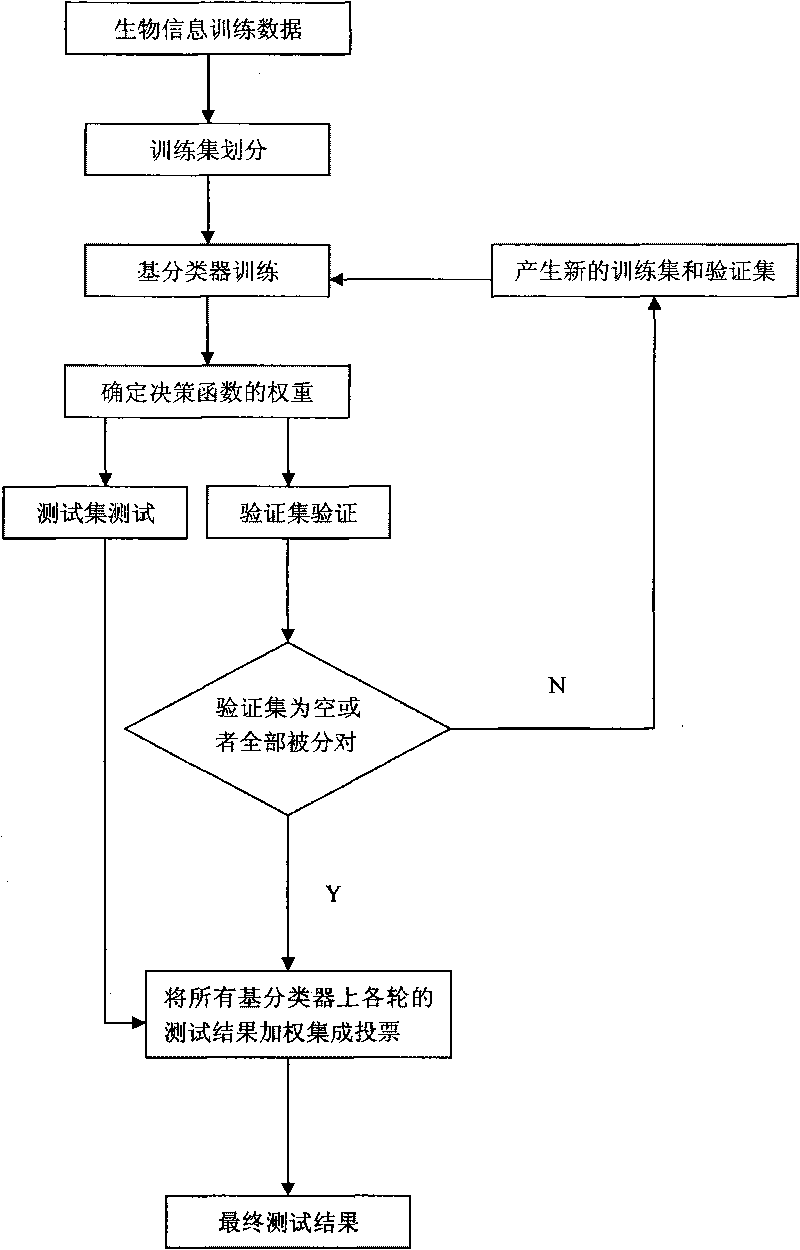
[0042] refer to figure 1 , the specific implementation process of the present invention is as follows:
[0043] Step 1. Determine the biological information data to be trained and tested.
[0044] This method is a bioinformatics data recognition problem, so first there are some training samples with labels. In the experiment, 40% of the labeled data is randomly selected as the training set X, and the other part is used as the test set.
[0045] Step 2. Normalize the determined training set data.
[0046] For the determined training set data, the following formula is used to normalize the data to remove the influence of the magnitude of the data, and to obtain the characteristics of the normalized training data:
[0047]
[0048] Among them, v=(f 1 ,, f 2 ,..., f n ) represents the training data, min(v) represents (f 1 ,, f 2 ,..., f n ) among the minimum value, max(v) means (f 1 ,, f 2 ,..., f n ) of the maximum value. Thus v'=(f' 1 ,, f 2 ',..., f n ') is the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com