Method for preprocessing and identifying electron transfer dissociation (ETD) mass spectrum
An electron transport and preprocessing technology, applied in the field of protein identification, can solve the problems of poor accuracy and low efficiency of ETD mass spectrometry data analysis, and achieve the effect of avoiding interference and improving the identification effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
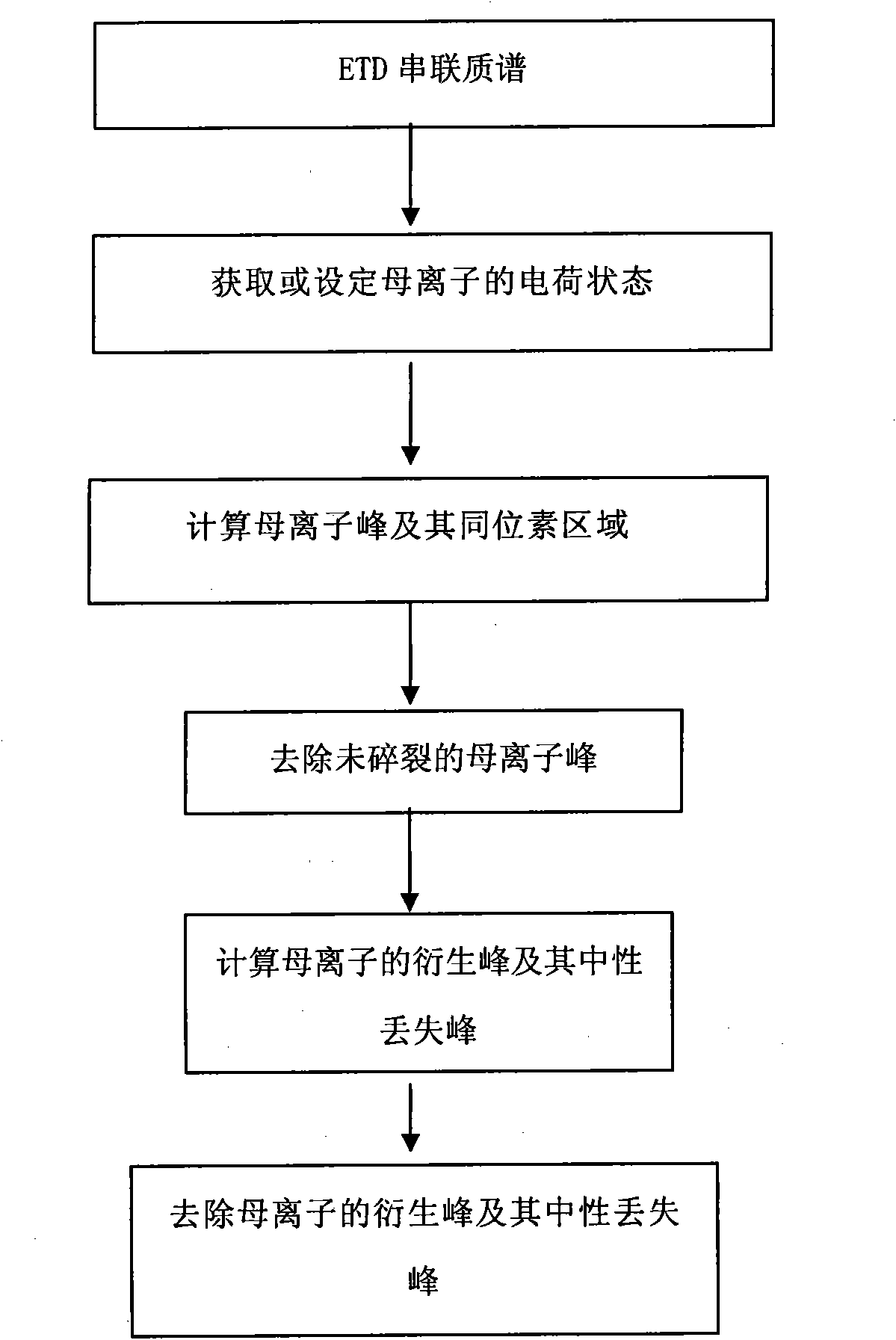
[0051] The present invention will be described below in conjunction with the accompanying drawings and specific embodiments.
[0052] Before the present invention is described in detail, in order to facilitate understanding, the meanings of several technical terms involved in the present invention are explained separately.
[0053] Precursor ion: Refers to fragmented peptide ions selected from the primary spectrum.
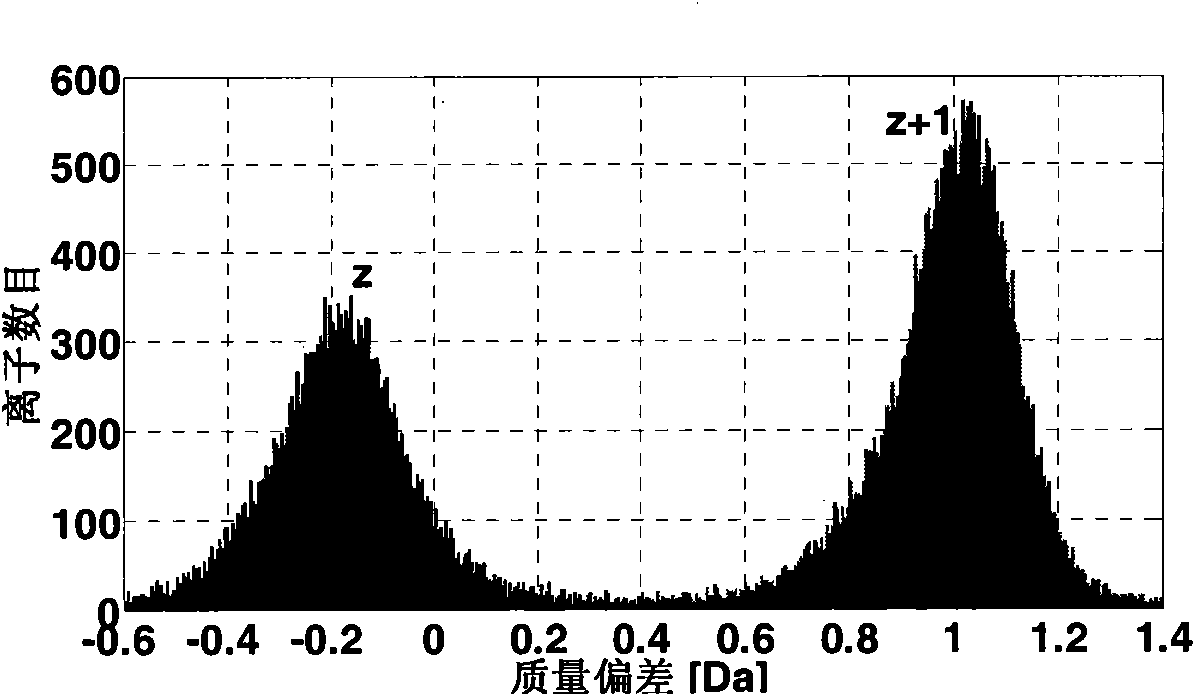
[0054] Derived peak: refers to the spectrum peak in which the protonated polypeptide ion interacts with electrons, and the mass is basically unchanged but the charge is reduced.
[0055] Neutral loss peak: refers to the spectral peak generated by the neutral loss of the side chain of the derivative peak.
[0056] After explaining some technical terms involved in the present invention, the ETD experimental tandem mass spectrum generated by the ETD method is analyzed below in conjunction with examples.
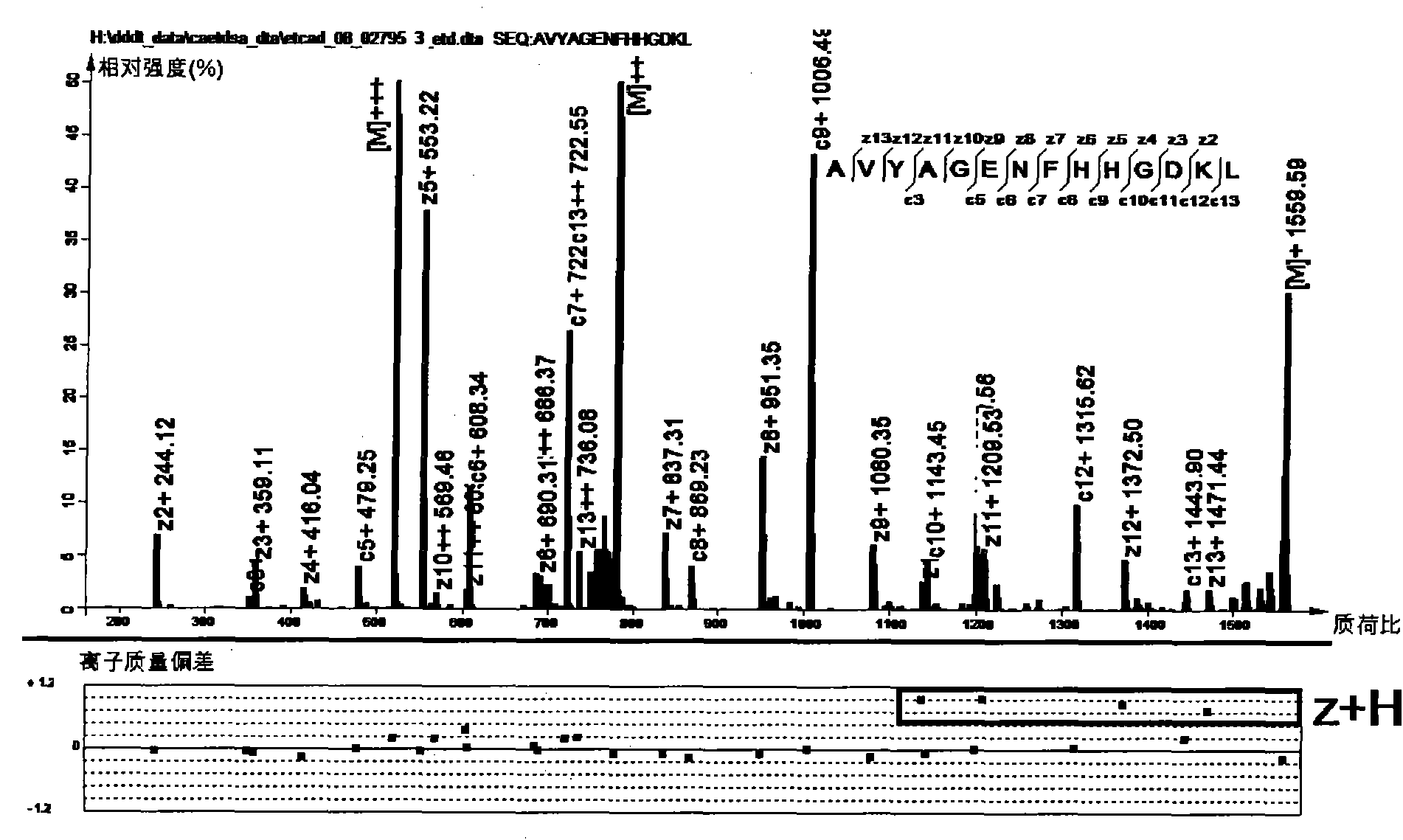
[0057] figure 1 An ETD spectrum of the polypeptide AVYAGENF...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com