Molecularly imprinted polymers for detecting microorganisms
A technology of molecular imprinting and polymers, applied in biological testing, analytical materials, measuring devices, etc., can solve problems that have not been used to detect or identify microorganisms
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
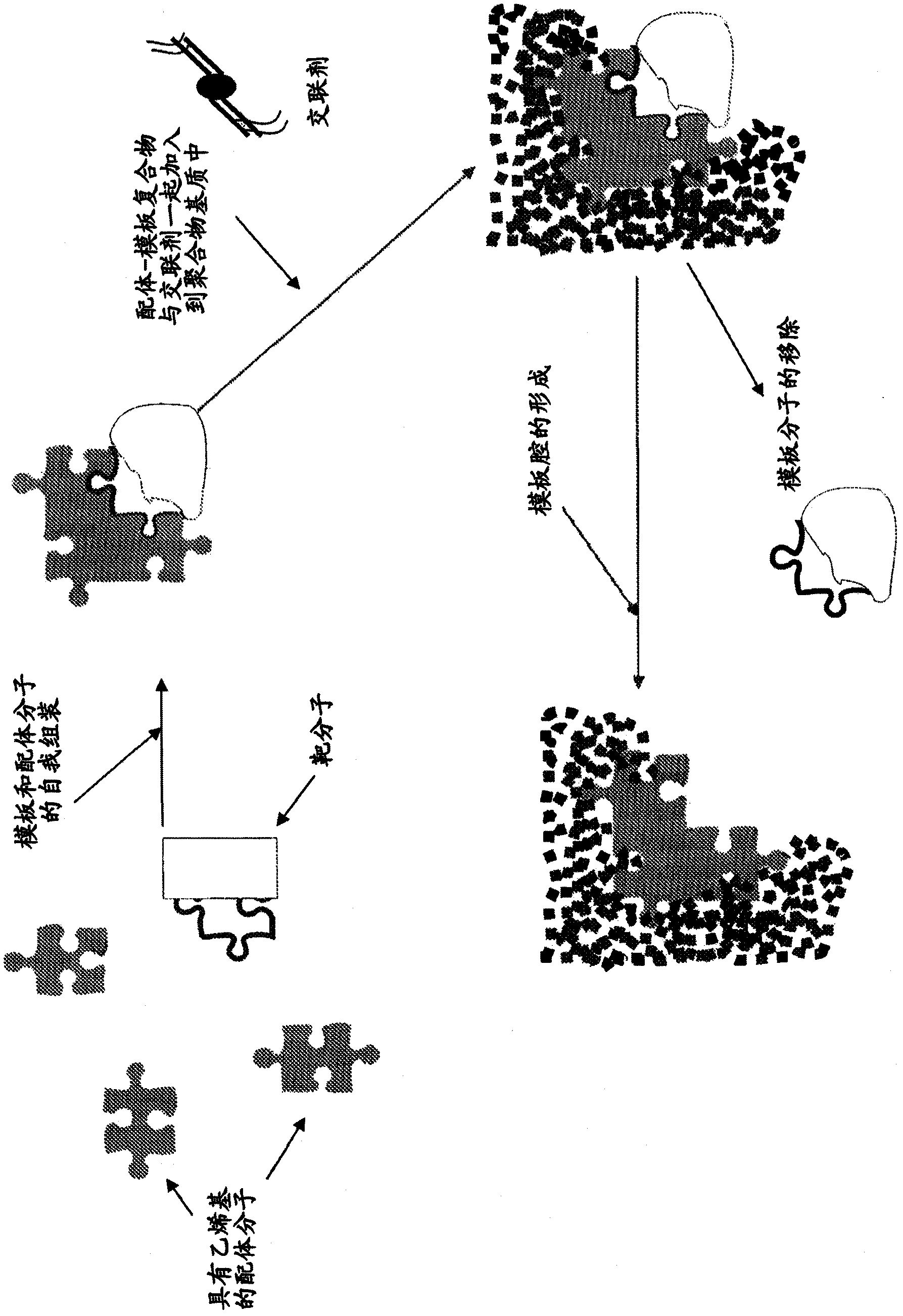
[0113] as in figure 1As demonstrated in , MIPs of the invention described herein can be made by generating a template for a portion of the target molecule and polymerizing functional monomers in the presence of this template. Functional monomers are able to bind to active sites on template molecules and then polymerize in the presence of excess cross-linking agents. While polymerization can be performed in the presence of template molecules, subsequent removal of the template molecules can leave cavities with a shape and arrangement of functional groups that are complementary to those of the template molecules. Thus, the resulting MIPs are capable of exhibiting the ability and selectivity to tightly rebind the template molecule.
Embodiment 2
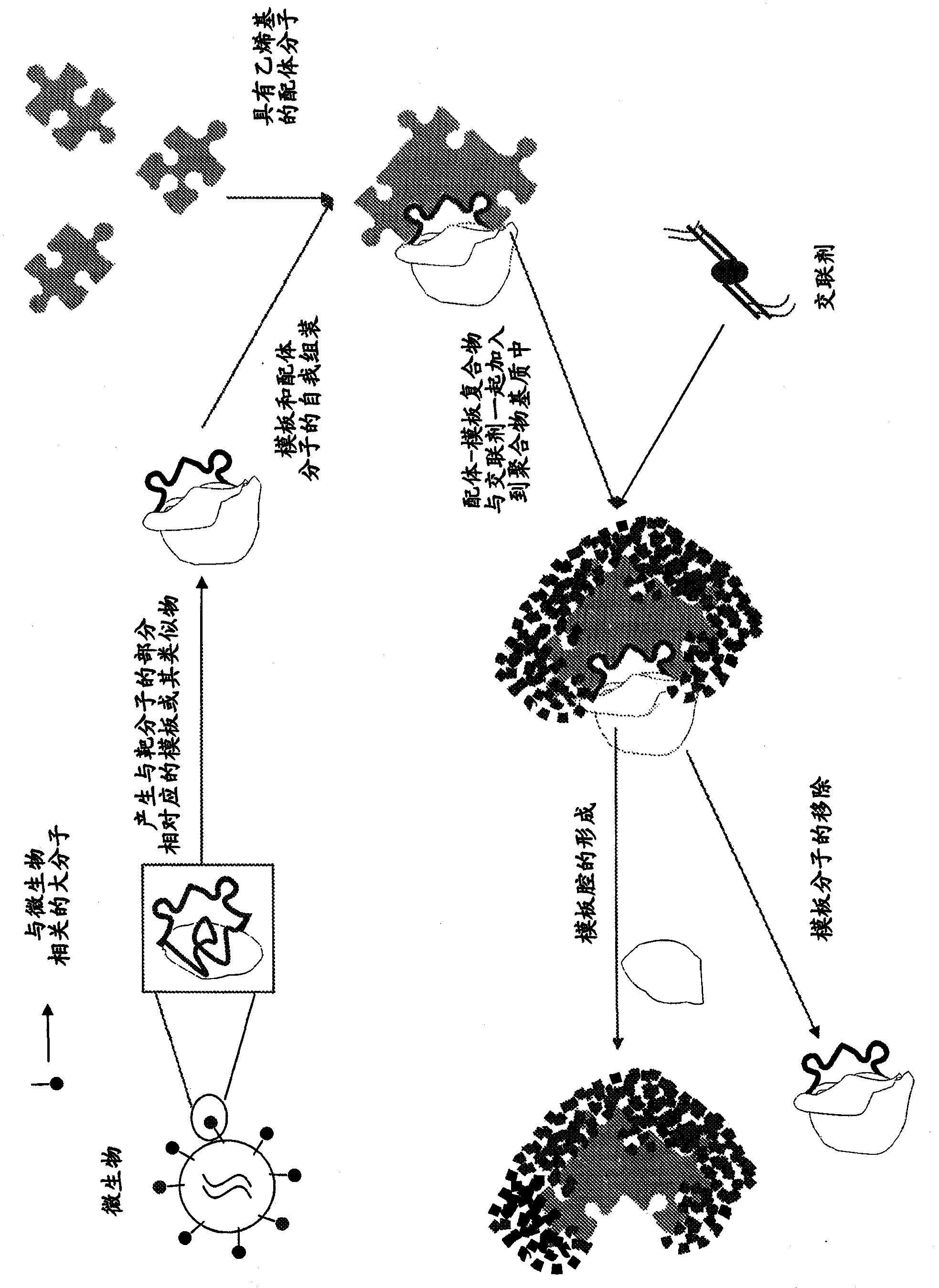
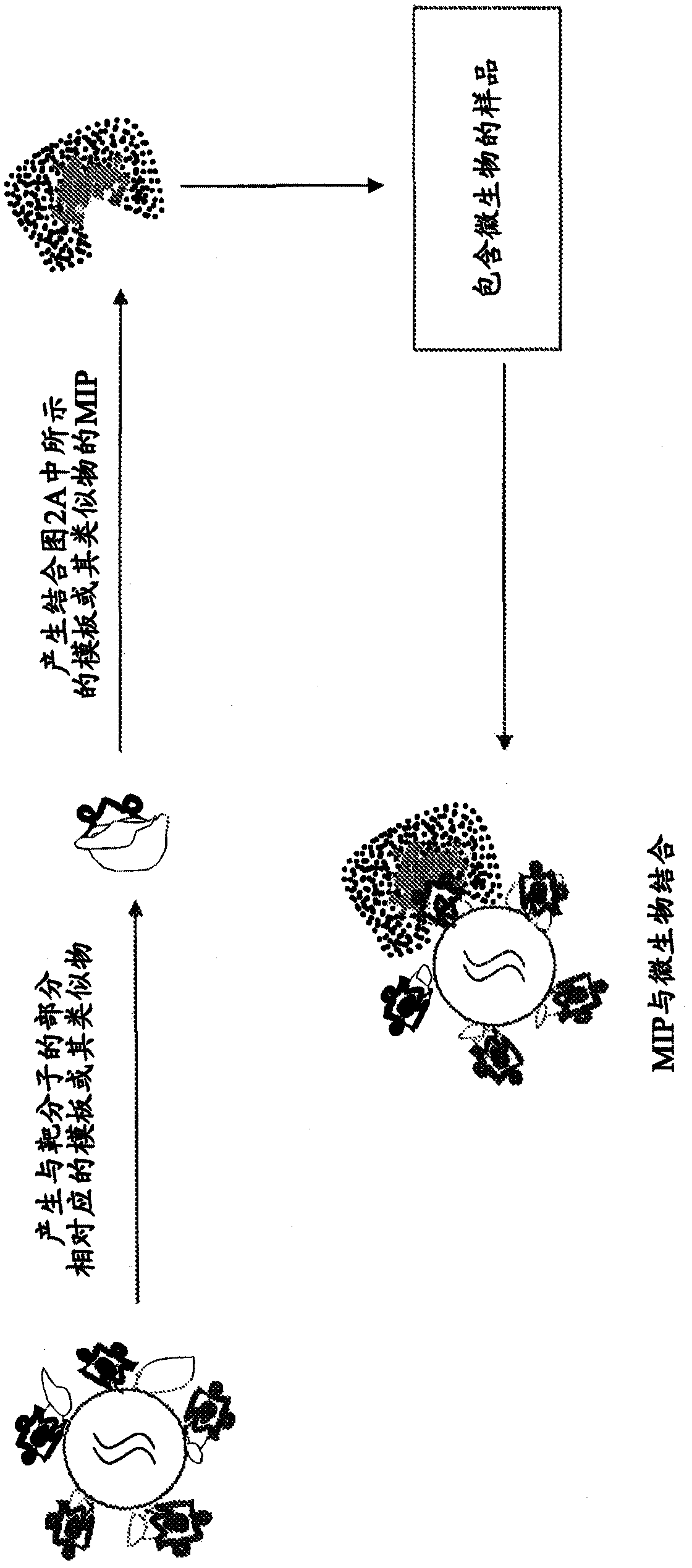
[0115] Figure 2A and 2B A schematic diagram showing the detection of bacteria using the MIP of the present invention. After identification of unique macromolecules associated with bacteria, templates for portions of macromolecules comprising one or more epitopes can be generated. Functional monomers can be polymerized in the presence of template molecules such that the monomers bind to active sites on the template molecules, and monomers can be further polymerized in the presence of excess cross-linking agents. Subsequent removal of the template molecule ( Figure 2A ) is able to leave a cavity with a shape and arrangement of functional groups complementary to those of the functional groups that are part of the macromolecules unique to the microbe. The resulting imprinted polymers can thus exhibit the ability and selectivity to tightly bind portions of macromolecules associated with microorganisms and identify said microorganisms ( Figure 2B ).
Embodiment 3
[0117] Using MIP to detect methicillin-resistant Staphylococcus aureus (MRSA)
[0118] One embodiment of the invention involves the detection of MRSA using the produced MIPs that bind PBP2A. In particular, MIPs can be generated using epitope amino acid sequences corresponding to different amino acid sequences within PBP2A as template molecules. For example, the template molecule can be designed to comprise the amino acid sequence (see Table 1 below) MKKIKIVPLILIVVVVGFGIYFYAS (SEQ ID NO: 1); KKIKIVPL (SEQ ID NO: 2); KIKIVPLI (SEQ ID NO: 3); QNWVQDDTF (SEQ ID NO : 4); KEYKGYKDDAVIGK (SEQ ID NO: 5); EYKGYKDD (SEQ ID NO: 6); YKGYKDDA (SEQ ID NO: 7); DKKEPLLNKFQITTS (SEQ ID NO: 8); KEPLLNKF (SEQ ID NO: 9); (SEQ ID NO: 10); PLLNKFQI (SEQ ID NO: 11); GYNVTRYEVVN (SEQ ID NO: 12); GVGEDIPSDYPFYNAQILD (SEQ ID NO: 13); DYPFYNAQ (SEQ ID NO: 14) and / or fragments thereof, They uniquely provide surface imprints with specificity for PBP2A. The MIP generated by using these amino acid sequen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com