PCR(Polymerase Chain Reaction) primers and method for identifying mycobacterium bovis
A technology of Mycobacterium bovis and Mycobacterium, which is applied in the field of PCR identification, can solve the problems of high economic cost and unfavorable clinical testing samples, achieve simple operation, high-throughput diagnosis and detection, and reduce false negative or false positive results. the effect of the probability of
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] The design of embodiment 1PCR specific primer
[0057] The 16S rDNA gene corresponding to 16SrRNA is highly conserved in the bacterial genome, and can identify the principle of bacteria of the genus. Firstly, primers are designed according to the 16Sr RNA conserved region of Mycobacterium. The target fragment size is 543bp sequence, which is designed using software and Primer 3 Primer, the primer sequence is:
[0058] 16SrRNA-F 5'-ACGGTGGGTACTAGGTGTGGGTTTC-3';
[0059] 16SrRNA-R 5'-TCTGCGATTAGCGACTAAGACTTCA-3';
[0060] Rv0577 is a restricted gene of MTBC, primers were designed according to this gene, and the size of the target fragment amplified by PCR was 786bp. The primer sequences are:
[0061] MTBC-F 5'-ATGCCCAAGAGAAGCGAATACAGGCAA-3';
[0062] MTBC-R 5'-CTATTGCTGCGGTGCGGGCTTCAA-3';
[0063] Since the deleted region 7 (deleted region7, RD7) does not exist in other MTBC strains, only Mycobacterium tuberculosis has this gene region, and primers are designed accor...
Embodiment 2
[0072] The establishment of embodiment 2PCR amplification method
[0073] 1. The PCR reaction system uses the total bacterial DNA as a template for PCR reaction. The 20 μL reaction system is:
[0074]
[0075] 2. PCR reaction conditions
[0076] The reaction program of PCR containing 16SrRNA, MTBC, PncA primer pair is: pre-denaturation at 94°C for 5 minutes, then denaturation at 94°C for 40s, annealing at 60°C for 40s, extension at 72°C for 40s, 31 cycles, and finally extension at 72°C for 10 minutes;
[0077] The reaction program of PCR containing MT primer pair is: pre-denaturation at 94°C for 5 minutes, then denaturation at 94°C for 40s, annealing at 61°C for 40s, extension at 72°C for 40s, 31 cycles, and finally extension at 72°C for 10 minutes;
[0078] The reaction program of PCR containing the RD1 primer pair was: pre-denaturation at 94°C for 5 min, followed by denaturation at 94°C for 40 s, annealing at 62°C for 40 s, extension at 72°C for 40 s, 31 cycles, and fina...
Embodiment 3
[0080] The specificity test of embodiment 3PCR method
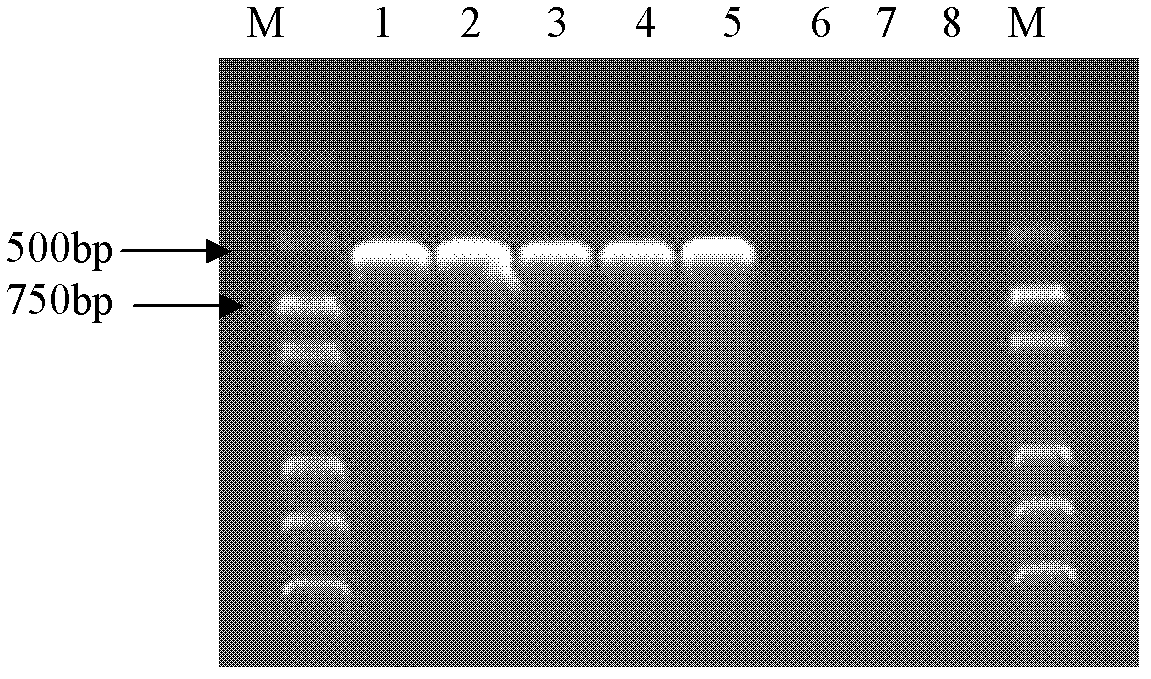
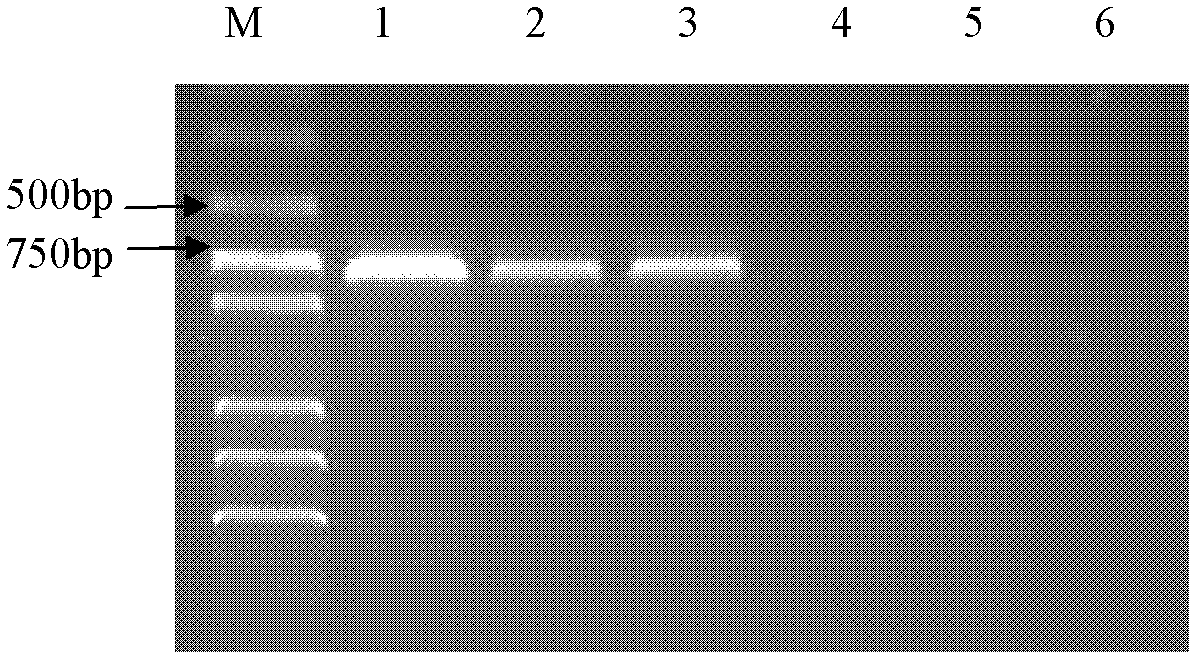
[0081]Mycobacterium tuberculosis C68503, Mycobacterium bovis C68001, Mycobacterium bovis BCGC68017, Mycobacterium smegmatis cm 2 155. The DNA of Mycobacterium avium C68201, Staphylococcus CAU0411, and Escherichia coli CAU0439 was used as a template, and water was used as a blank control. 5 pairs of primers of the present invention were used respectively, and the PCR method established by the present invention was used for inspection. After the PCR reaction ended, the agar Judgment results of specific amplification bands by sugar gel electrophoresis.
[0082] test results:
[0083] PCR results of 16SrRNA primers: the PCR amplification products of Mycobacterium were positively amplified, while the other strains and the blank control had no target amplified fragments. For the results, see figure 1 .
[0084] PCR results of MTBC primers: positive amplification of Mycobacterium tuberculosis, Mycobacterium bovis and BCG, bu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com