DNA (deoxyribonucleic acid) library and preparation method thereof, as well as DNA sequencing method and device
A DNA library and DNA sequencing technology, applied in the field of molecular biology, can solve problems such as difficulty, difficulty in ensuring success rate, and inability to obtain fosmid clone ends, and achieve the effect of improving availability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Example 1: DNA library construction and sequencing of the penguin genome
[0070] 1. Construction of DNA library of penguin genome
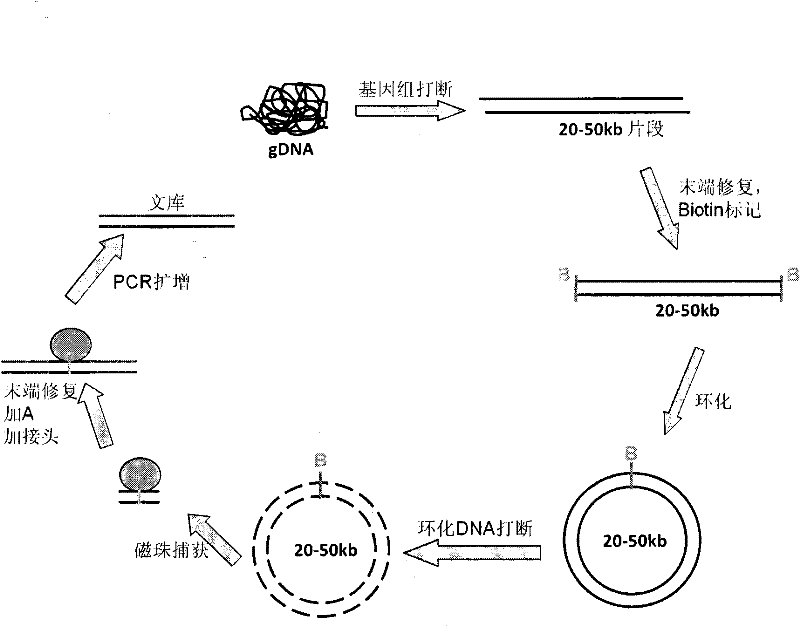
[0071] 1) Random interruption of sample genomic DNA
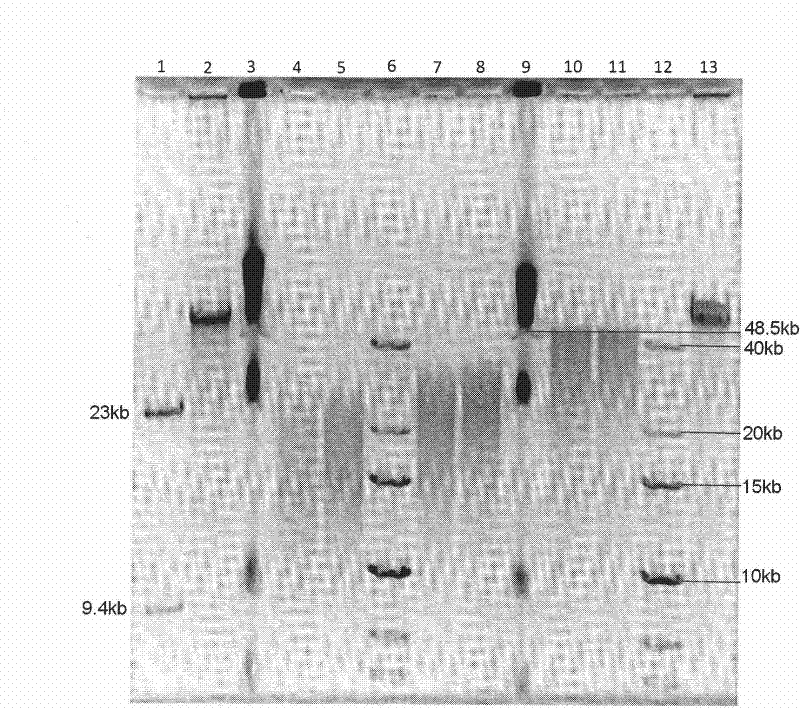
[0072] Genomic DNA of Adrien penguin (Pygoscelis adeliae) was used as the library construction sample, and an end-paired library with an insert fragment of 40-45kb was constructed at a starting point of 50 μg, and a standard Hydroshear instrument (GeneMachine, San Carlos, CA., USA) was used for fragmentation , set the interruption parameters to speed (speed code) 15, cycle number (cycles) 30, and interruption reaction system to 100 μl.
[0073] After fragmentation was completed, it was recovered into EP tubes, and the fragmented DNA fragments were purified using Agencourt AMPure Beads (BECKMAN COULTER). Add 1.8 times the volume of Agencourt AMPure Beads to the fragmentation reaction system, mix them upside down, and place them at room temperature for 10 Minutes to fully combine the ...
Embodiment 2
[0093] Example 2: DNA library construction and sequencing of plum blossom genome
[0094] The DNA library construction and sequencing of the Prunus mume genome were performed in the same manner as in Example 1, except that the genomic DNA sample used was Prunus mume genomic DNA. The DNA library (40kb paired-end DNA library) sequence result of the Meihua genome was obtained.
[0095] Sequencing Results and Analysis
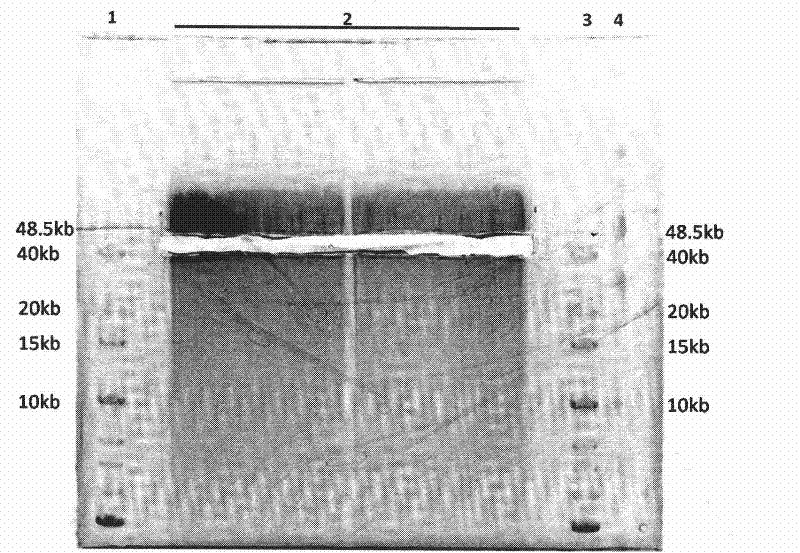
[0096] Sequence the paired-end DNA library of plum blossoms obtained on the Illumina HiSeq 2000 sequencing platform to obtain paired-end sequence information with an insert fragment of 40kb. These data were used for genome assembly of plum blossoms, and the data were compared to the genome of plum blossoms using SOAPdenovo software In terms of sequence, it was verified that the distance span of the paired-end sequence obtained by sequencing the library was 40kb, which was in line with the expected fragment range ( Figure 5 ). Use SOAPdenovo software to assemb...
Embodiment 3
[0097] Example 3: DNA library construction and sequencing of human genome
[0098] The DNA library construction and sequencing of the Meihua genome were performed in the same manner as in Example 1, except that the genomic DNA sample used was human genomic DNA. The DNA library (40kb paired-end DNA library) sequence result of the human genome was obtained.
[0099] Sequencing Results and Analysis
[0100] The obtained human paired-end DNA library was sequenced on the Illumina HiSeq 2000 sequencing platform, and the paired-end sequence information with an insert fragment of 40kb was obtained. These data were used for human genome assembly, and the data were compared to the human genome using SOAPdenovo software In terms of sequence, it was verified that the distance span of the paired-end sequence obtained by sequencing the library was 40kb, which was in line with the expected fragment range ( Figure 6 ). Use SOAPdenovo software to assemble the human genome. When the human...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com