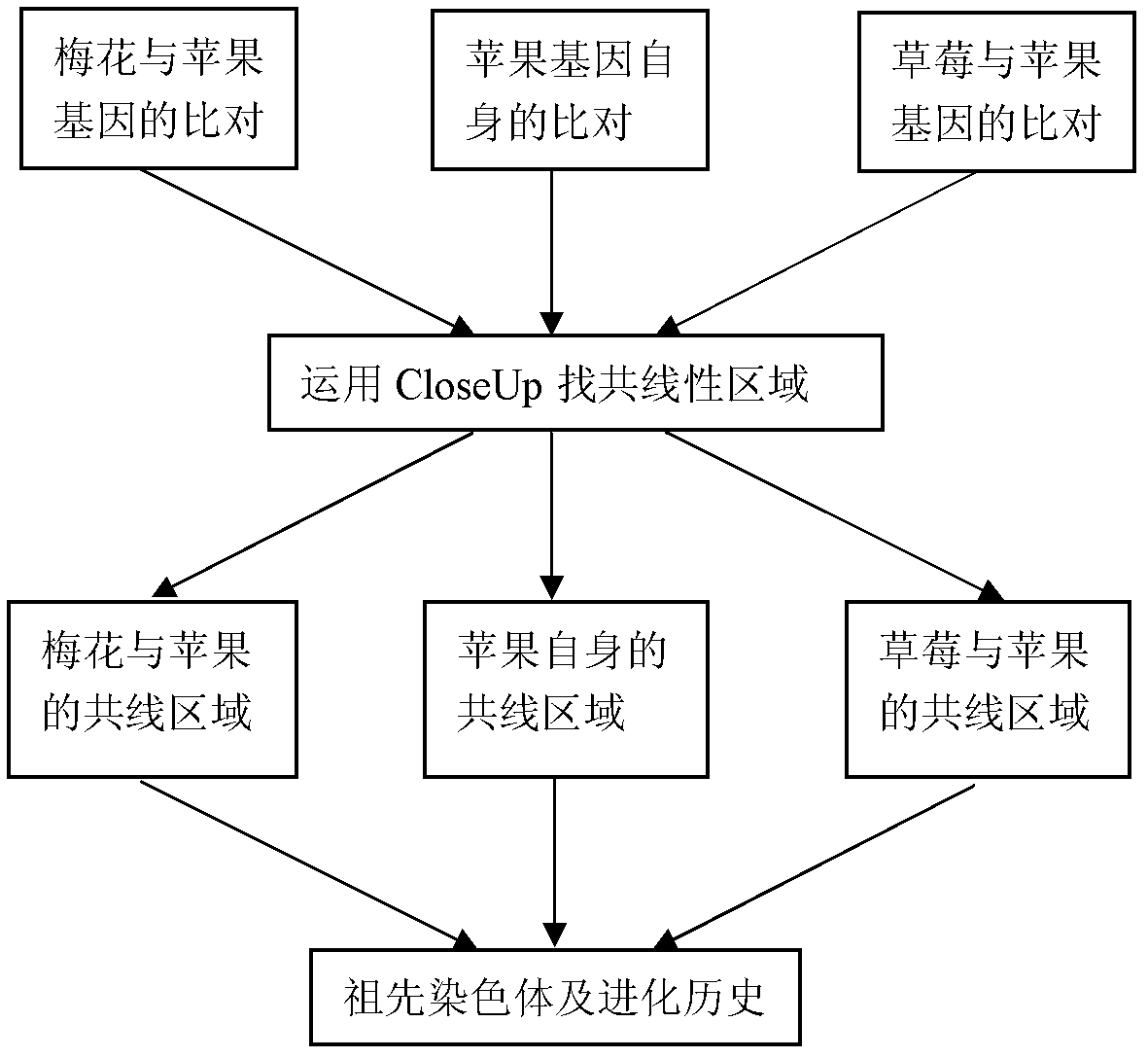
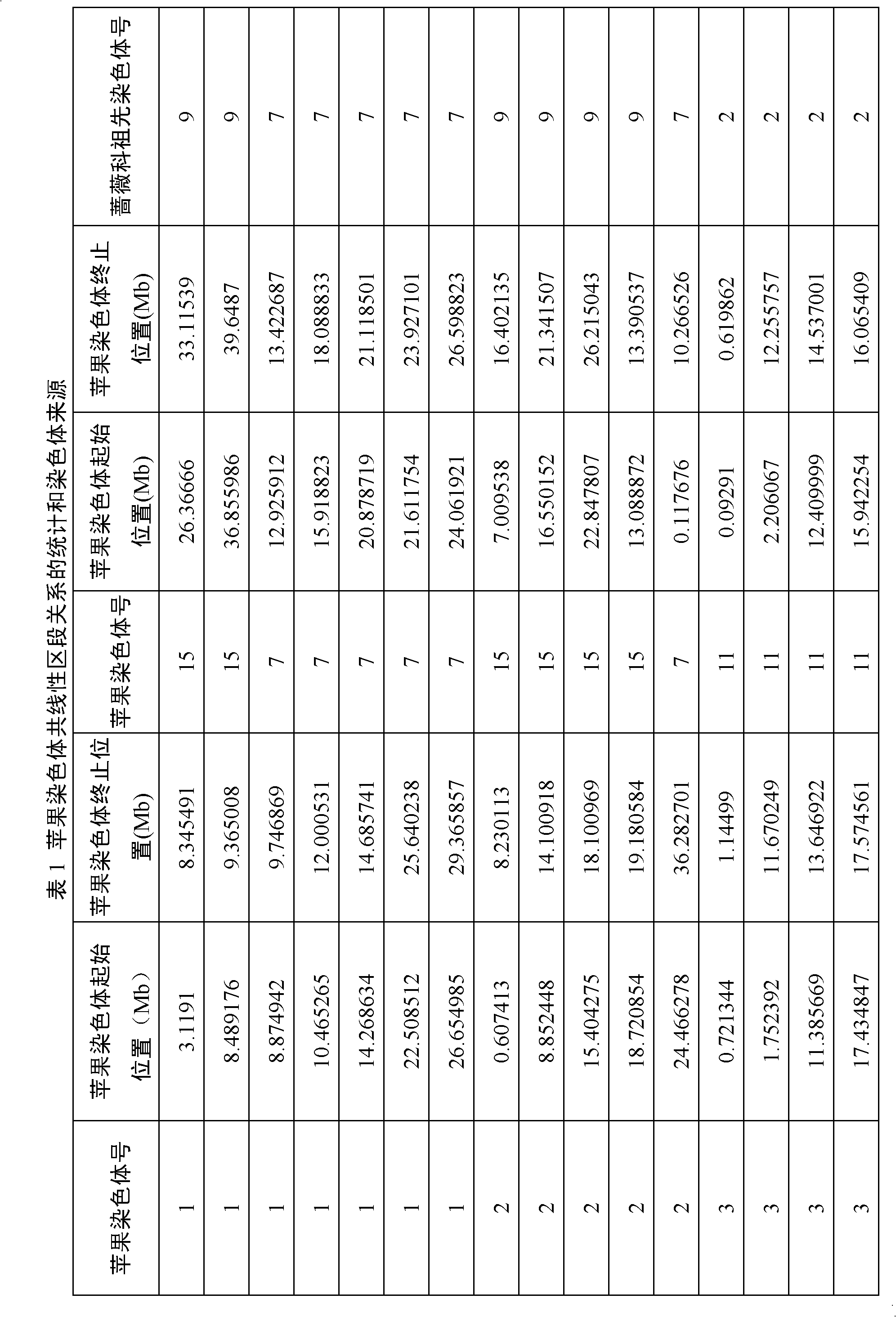
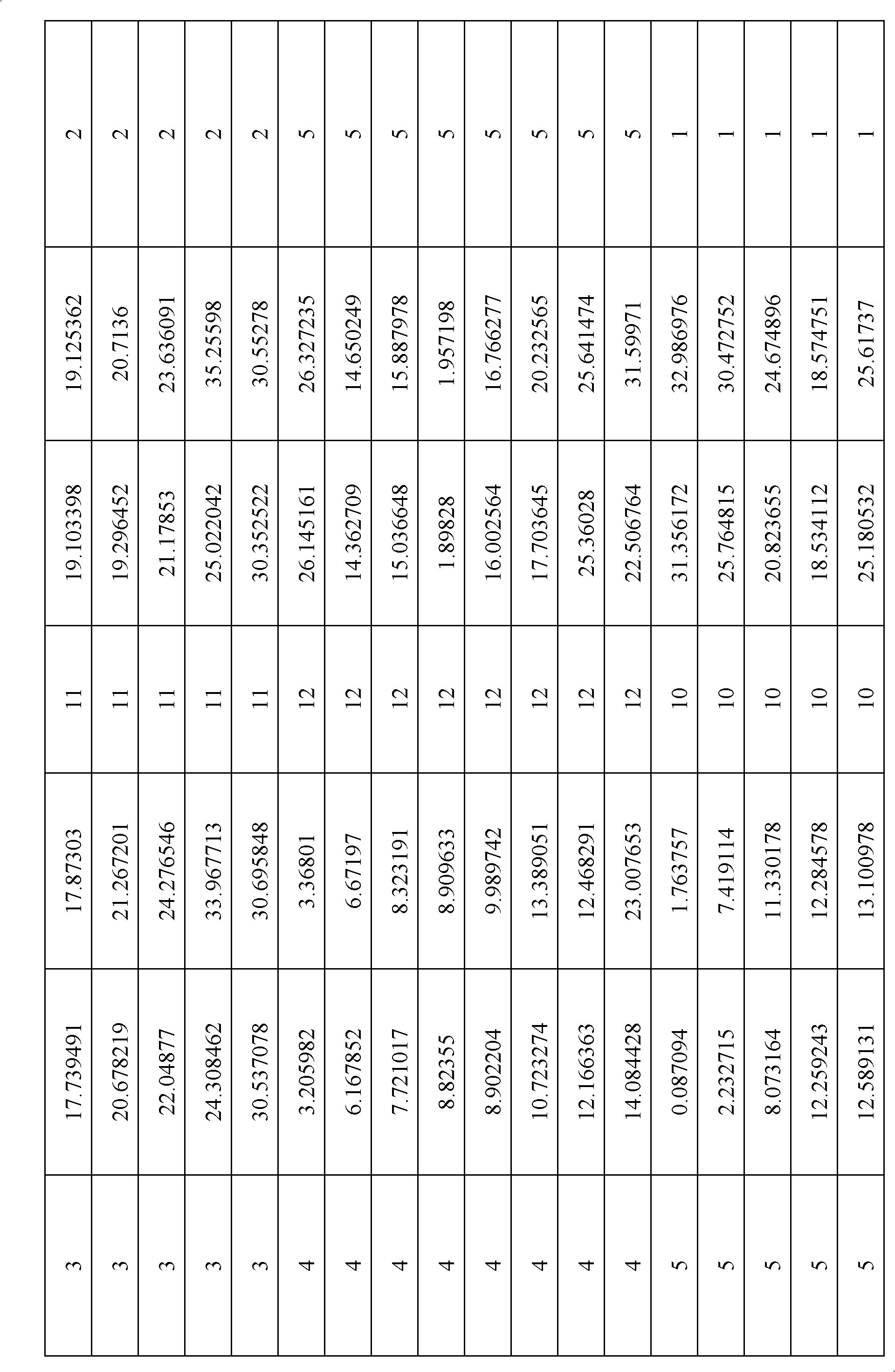
Method for constructing rosaceae original chromosome
A chromosome and Rosaceae technology, applied in the field of bioinformatics to avoid interference and expand homologous regions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] 1. Collection of genome data of apple, strawberry and plum blossom. from http: / / www.rosaceae.org / projects / apple_genome Obtain the apple gene CDS sequence and gene location information on the chromosome; download the strawberry gene CDS sequence and gene location information on the chromosome from http: / / www.strawberrygenome.org / ; positioning information. 2. Identification of homologous genes. Cumulative identity percentage (CIP: cumulative identity percentage) and cumulative alignment length percentage (CALP: cumulative alignment length percentage) of two gene alignments. It is calculated according to the comparison result of the comparison species (query) gene and the reference species (subject) gene blastn and the length of the comparison gene. Compare the CIP of gene A and reference gene B=∑(N AB / L AB )×100, where N AB is the length of each alignment between gene A and gene B, L AB is the length of gene A and gene B participating in the alignment; CALP of ge...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com