Key protein predicating method based on priori knowledge and network topology characteristics
A key protein and network topology technology, applied in the field of predicting new key proteins based on some known key proteins and biological network topological properties, can solve the problems of high cost and time-consuming chemical experimental methods, and achieve high cost and time-consuming solutions. , to achieve a simple effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
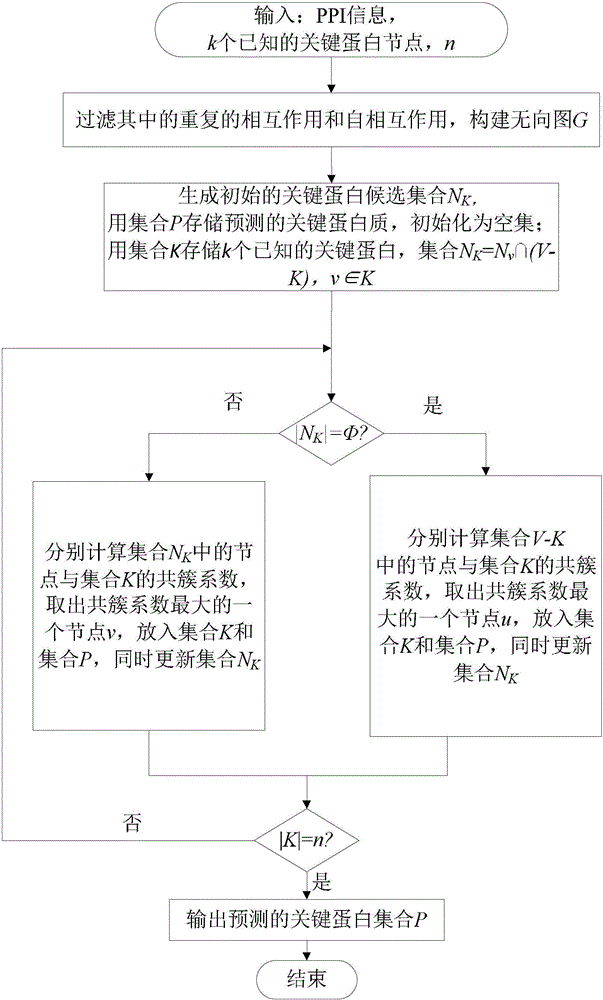
[0023] Specific embodiments of the present invention will be described in detail below in conjunction with the accompanying drawings.
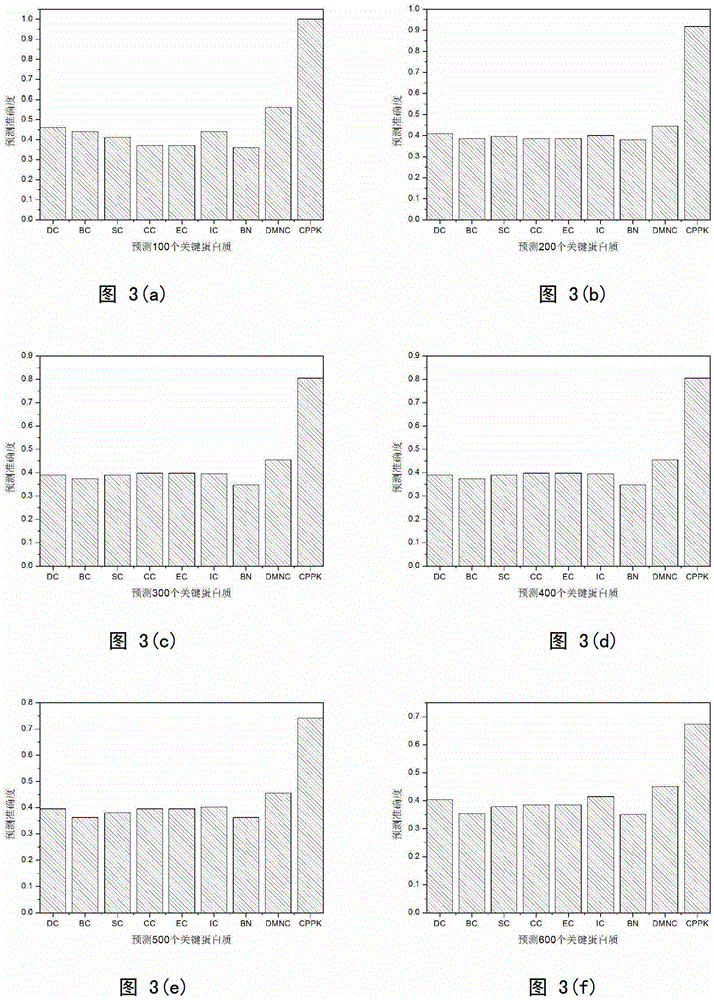
[0024]The most widely studied species is yeast, and there are already a certain number of key yeast proteins determined by experimental means. The present invention downloads the yeast protein interaction network data from the DIP (the database of interacting proteins) database. The interaction data were removed from self-interactions and redundant interactions, and the final protein interaction network consisted of 5093 yeast proteins and 24743 pairs of interactions. The key protein data used in the experiment comes from MIPS (Munich Informationcenter for Protein Sequences), SGD (Saccharomyces Genome Database), DEG (Database of Essential Genes) and SGDP (Saccharomyces Genome Deletion Project) four databases. Through comparison, among the 5093 proteins in the yeast PPI network used in the experiment, a total of 1167 are key proteins, 3591 are...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com