Porcine parvovirus VP2 affinity peptide and coding nucleotide thereof
A parvovirus and nucleotide technology, applied in the biological field, to achieve the effects of strong penetration, easy penetration into tissues, and short half-life
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Construction of Recombinant Expression Plasmid pET32a-VP2
[0033] (1) Amplification of VP2 sequence
[0034] (a) Design primers VP2-3 and VP2-4 according to the codons favored by bacteria
[0035] VP2-3: 5'-GG GGATCC ATGAGTGAAAATGTGGAA-3' (the dashed line indicates that the introduced enzyme cutting site is BamHI)
[0036] VP2-4: 5'-CCCC GTC GAC TTAGTATAATTTTCTTGG-3' (the dashed line indicates that the introduced enzyme cutting site is SalI)
[0037] (b) Perform conventional chain polymerase reaction to amplify the VP2 gene
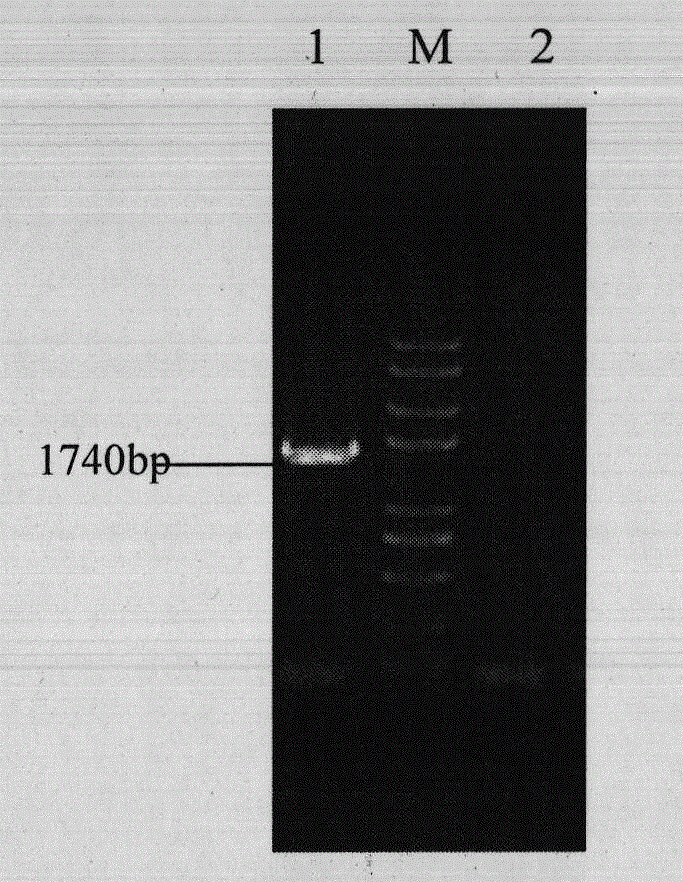
[0038] Use VP2-3 and VP2-4 as primers, VP2-T-6 as template, and perform PCR with ExTaq polymerase. PCR reaction conditions for VP2: 50 μL of the PCR reaction system contained in 50 μL system, including 5 μL of 10×EasyTaq Buffer , 4 μL dNTP, 1 μL VP2-3, 1 μL VP2-4, 0.5 μL VP2-T-6, 0.5 μL ExTaq polymerase, add sterile water to make up to 50 μL. The reaction program of the mixed solution in the PCR detection is: pre-denaturation at 95°C for 5...
Embodiment 2
[0041] Example 2 PCR and Enzyme Digestion Identification of Recombinant Expression Plasmid pET32a-VP2
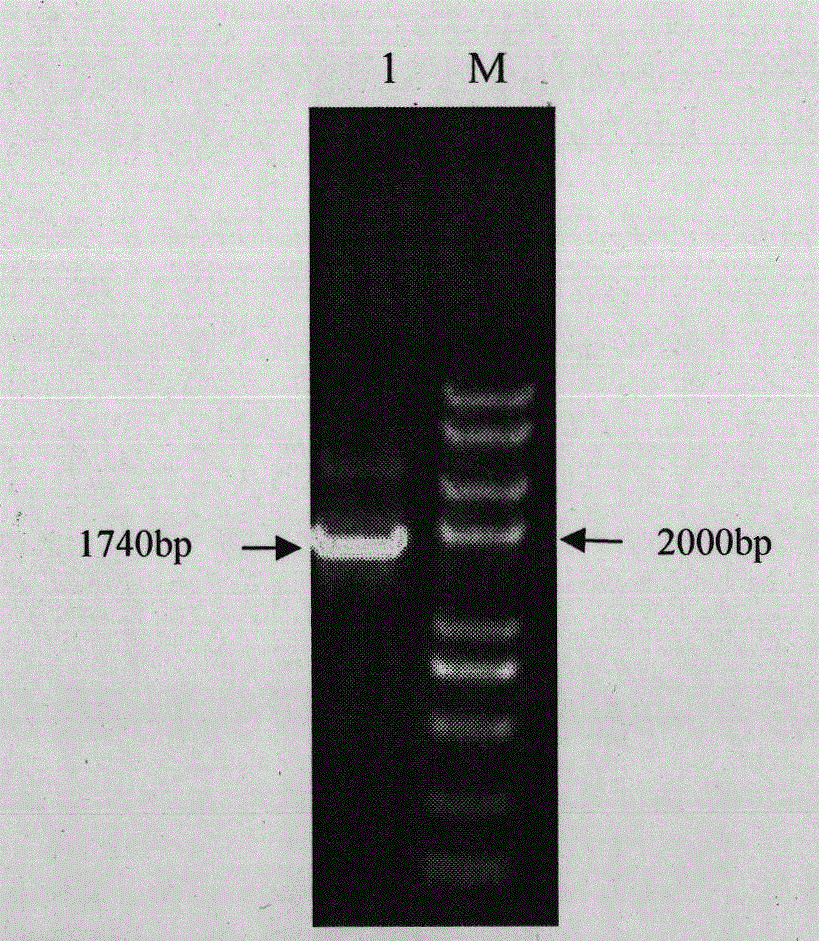
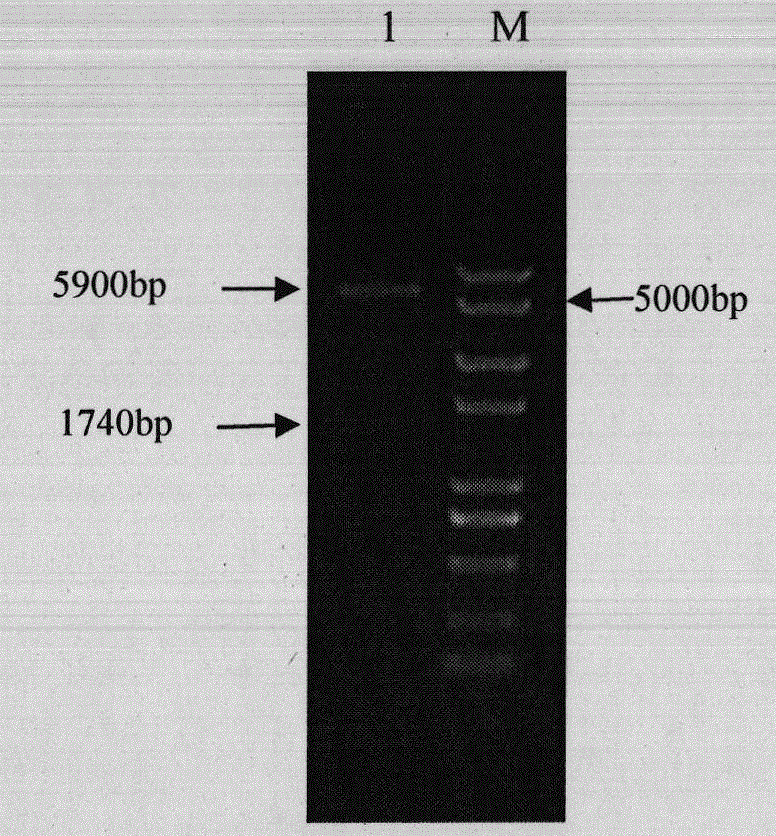
[0042] Select positive clones, extract plasmid pET32a-VP2, use VP2-3 and VP2-4 as primers for PCR identification, and perform double enzyme digestion with BamH I and Sal I, and detect the product by 1% agarose gel electrophoresis, and its position and prediction unanimous. The result is as figure 2 , indicating that the VP2 gene was successfully connected into the pET32a vector.
Embodiment 3
[0043] SDS-PAGE analysis of embodiment three induced expression products
[0044] The correctly identified recombinant plasmid pET32a-VP2 was transformed into E.coli Rosetta (DE3) competent cells, and IPTG was added to make the final concentration 0.6mmol / L, and cultured with shaking at 37°C. The test samples were processed and analyzed by SDS-PAGE electrophoresis. The result is as Figure 4 .
PUM
| Property | Measurement | Unit |
|---|---|---|
| control rate | aaaaa | aaaaa |
| control rate | aaaaa | aaaaa |
| control rate | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com