Protein-ligand binding site predicting method based on inquiry drive
A binding site and prediction method technology, applied in the field of bioinformatics protein-ligand interaction, can solve the problems of fitting over-optimization, low scalability, low usability, etc., to prevent over-optimization and over-fitting, The effect of improving prediction accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
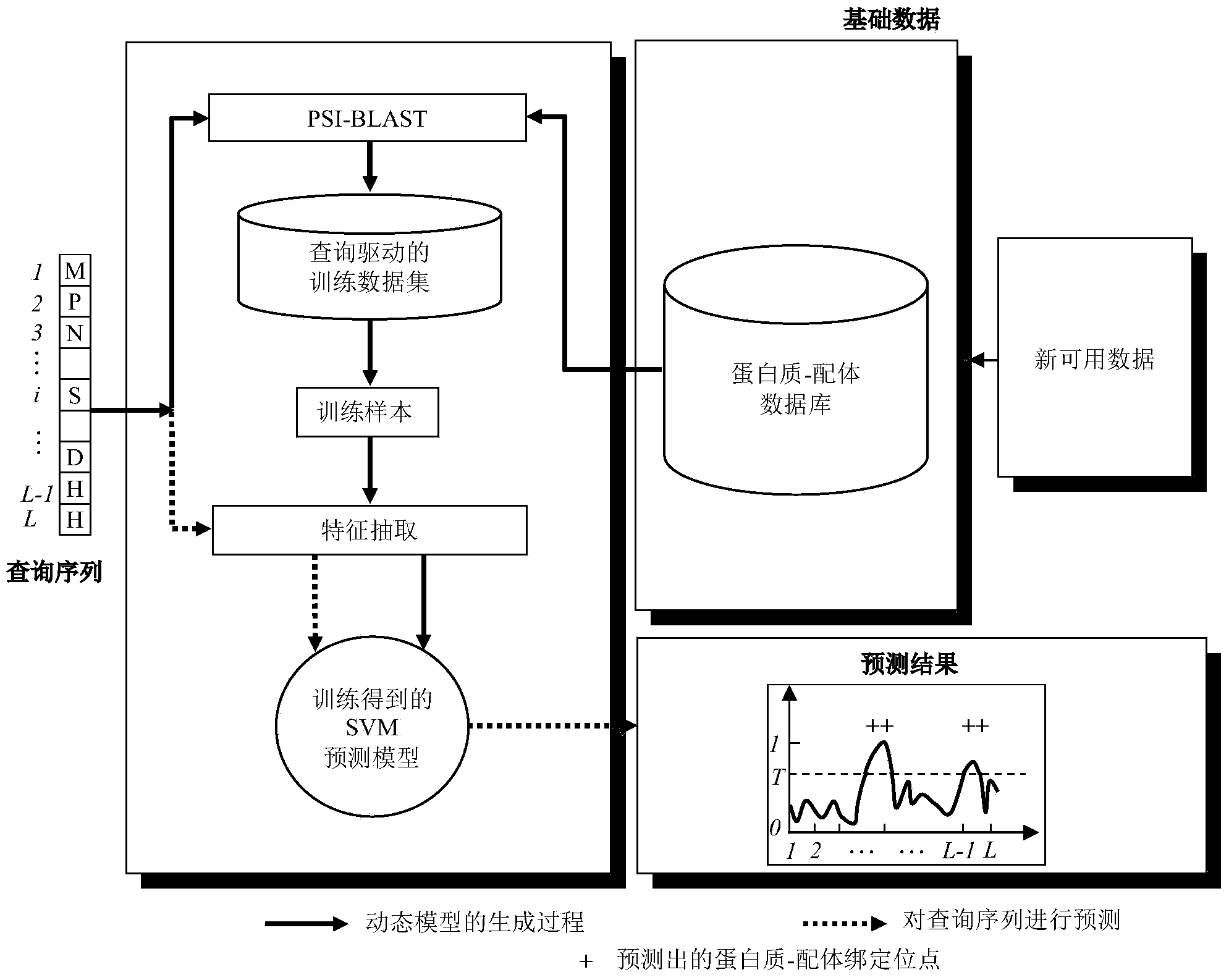
[0025] Such as figure 1 As shown, according to a preferred embodiment of the present invention, a query-driven protein-ligand binding site prediction method is used for a protein sequence to be predicted / queried (hereinafter referred to as a given query input q) Prediction is divided into two stages, namely, the dynamic model construction stage and the prediction stage, which are combined below figure 1 As shown, the implementation of the above two stages is described in detail.
[0026] (1) Dynamic model construction stage
[0027] The first step, using the PSI-BLAST tool software from the available data set D, namely figure 1 in the protein-ligand database lookup with a given query input q (i.e. figure 1 The query sequence in ) protein sequences with high homology constitute a query-driven and small-scale training data set D q-specific , so dynamically get a query-driven training data set, expressed as:
[0028] D. q-specific ←PSI-BLAST(q,D).
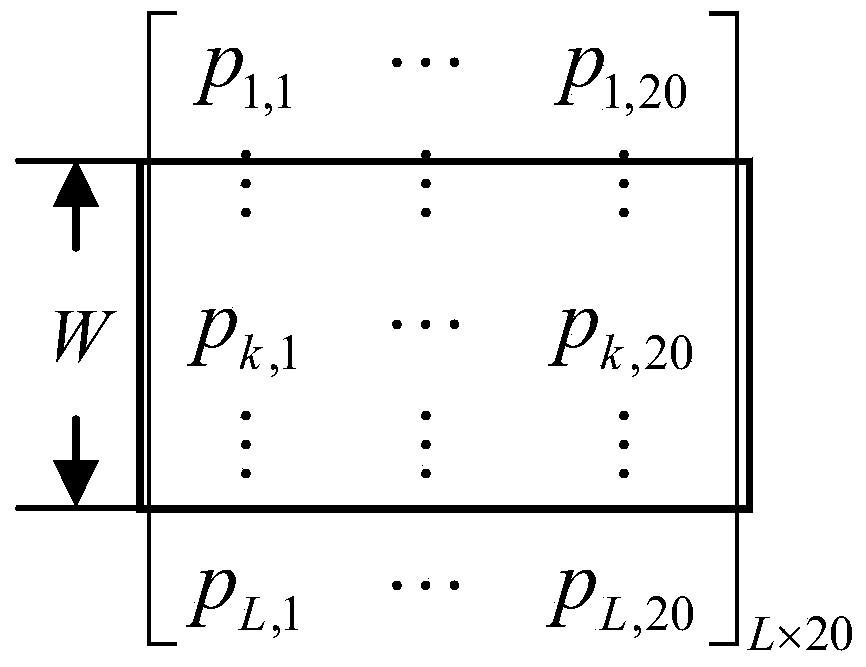
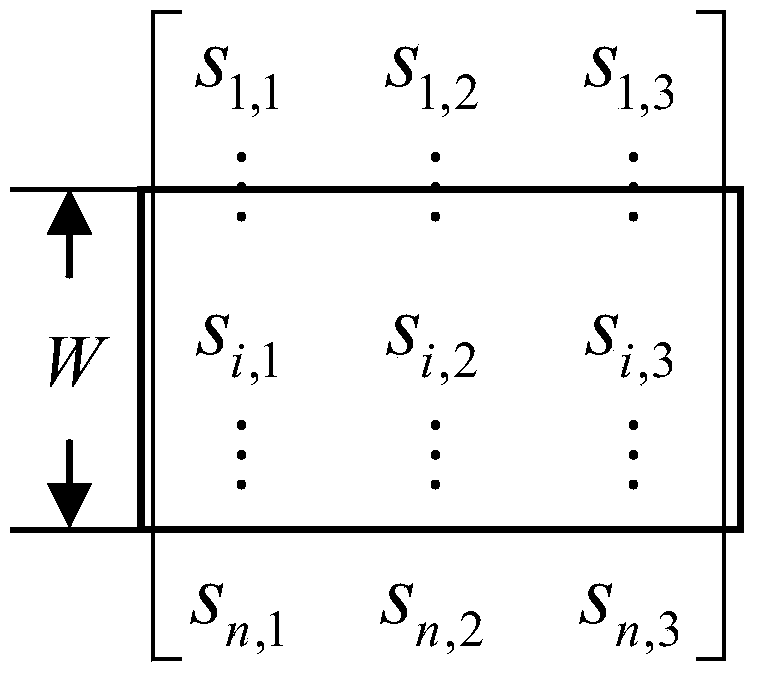
[0029] Such as figure ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com