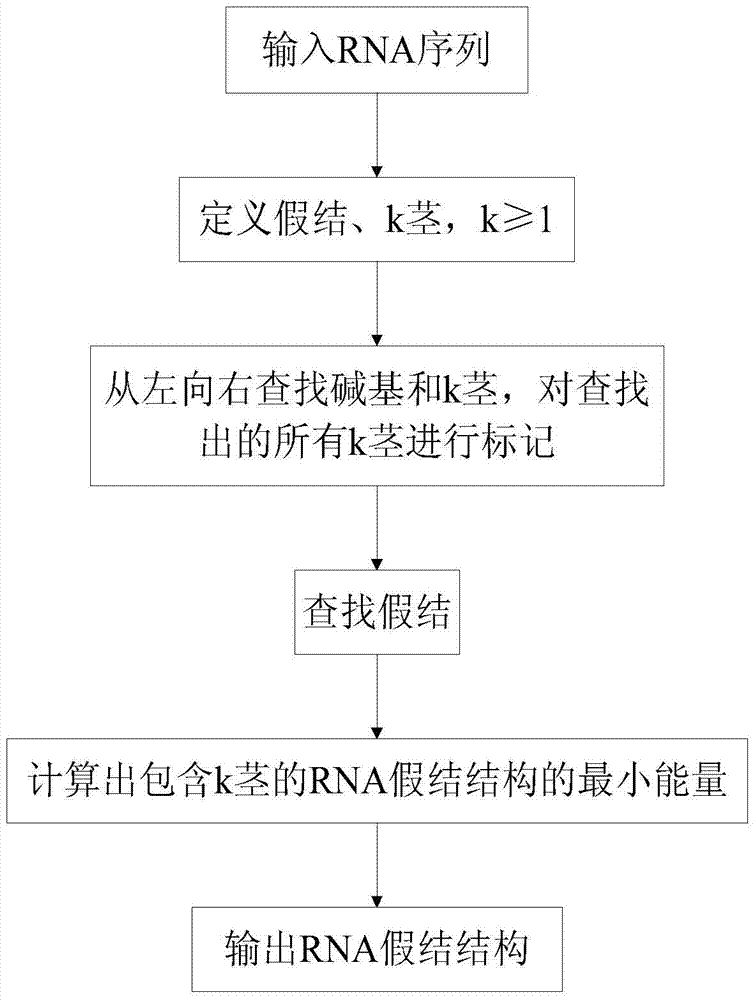
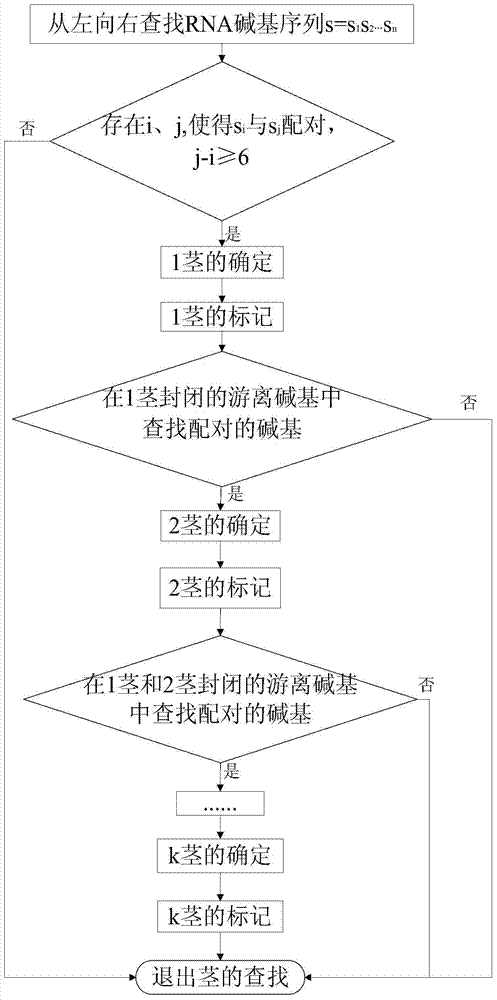
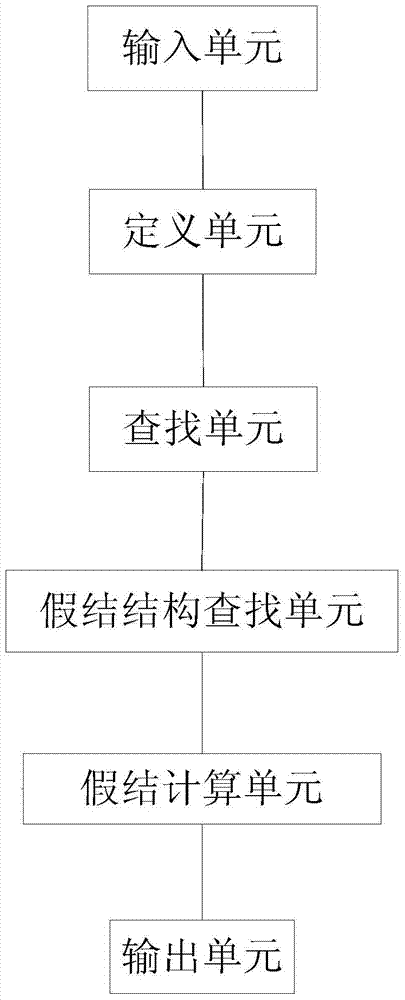
Method and device for predicting ribonucleic acid pseudoknot structure based on k-stem
A technology of ribonucleic acid and prediction method, applied in the field of bioinformatics engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] In RNA pseudoknot structure prediction, if k=1 or k=2 in k stems, the program calculations related to 1 stem and 2 stems are as follows.
[0043] Procedure 1: Calculation of energy and length for 1 stem and 2 stem
[0044]
[0045]
[0046] Figure 4 Give a simple false knot. Use two 1 stem (S 1 [1,19], S 1 [7,30]) and three subsequences (s 6,6 , s 13,14 , s 20,24 ) form a false knot. Since each 1-stem is determined by two parameters, the storage of 1-stem requires O(n 2 ) space, so the time complexity of calculating the false knot is O(n 4 ), the space complexity is O(n 2 ).
[0047] Depend on Figure 4 Know: W(1,30)=ES 1 (1,19)+ES 1 (7,30)+W(6,6)+W(13,14)+W(20,24)
Embodiment 2
[0049] Given a sequence s=s 1 the s 2 …s n , the sequence fragment s i,j =s i …s j , 1i,j The corresponding minimum energy of the secondary structure S containing pseudoknots. Let V(i,j) be s i and s j In the case of constituting a base pair (i,j), the subsequence S i,j The corresponding minimum energy of the secondary structure S containing pseudoknots.
[0050] Figure 5 Give the calculation schema of W(i,j) and V(i,j). W(i,j) containing pseudoknot structure is calculated by the following 4 cases:
[0051] 1)s j is an unpaired base, base s i and s j-1 The pairing relationship is not determined, such as Figure 5 .1, calculated W(i,j)=W(i,j-1);
[0052] 2)s i is an unpaired base, base s i+1 and s j The pairing relationship is not determined, such as Figure 5 .2, Calculated W(i,j)=W(i+1,j);
[0053] 3)s i and s k ,s k+1 and s j do not constitute a base pair and are in different subsequences S i,k and S k+1,j In the corresponding secondary structure, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com