Protease substrate screening method based on solid-loaded mixed protein as screening database
A screening method, protease technology, applied in biochemical equipment and methods, immobilized on/in organic carriers, determination/inspection of microorganisms, etc. Simple, specific and reliable results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
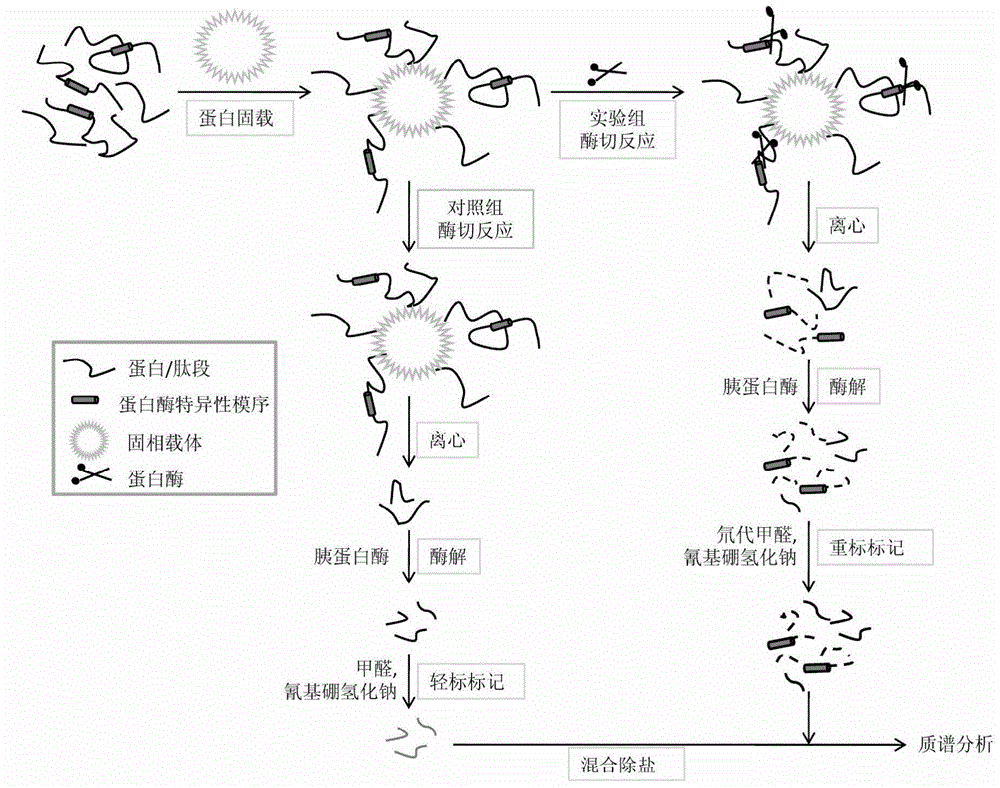
[0033] Example 1 Screening of caspase 3 substrates based on agarose microspheres as a carrier.
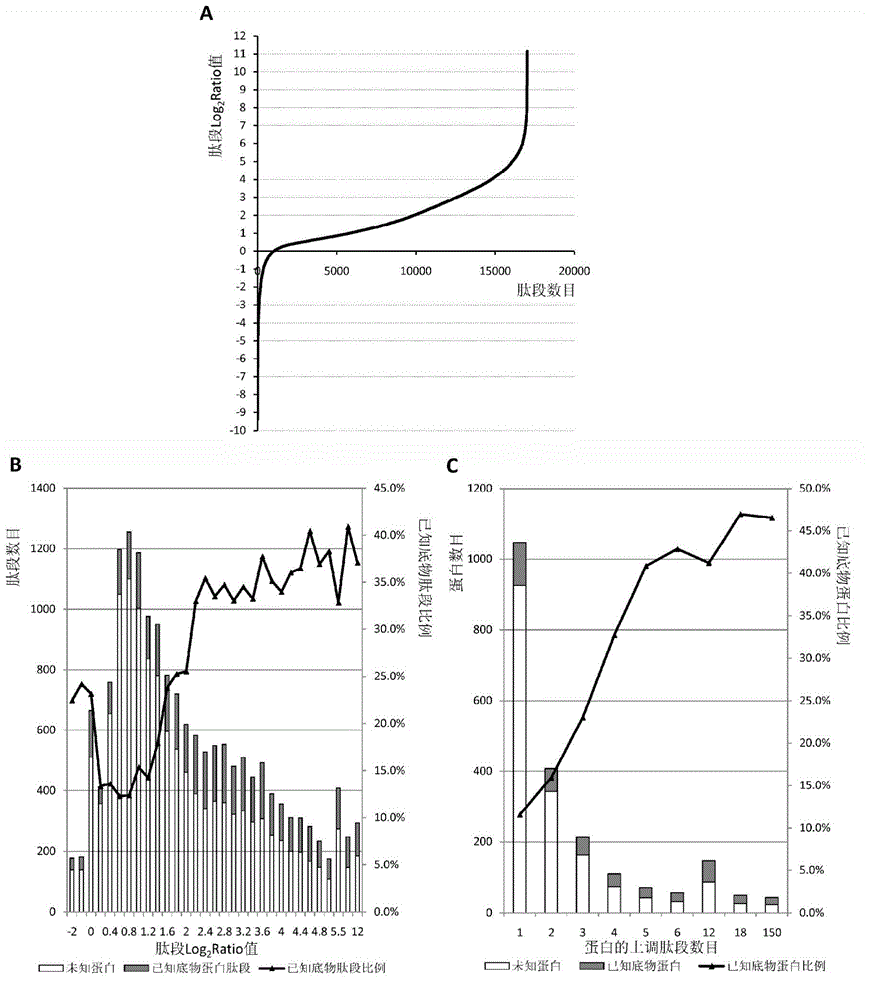
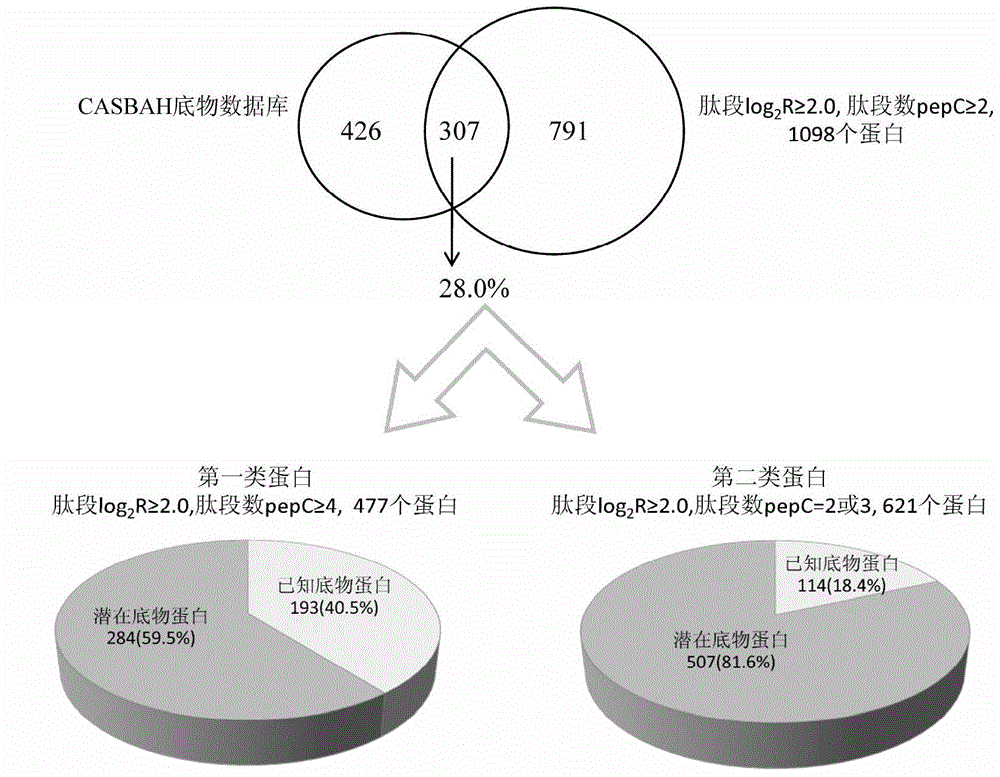
[0034] Preparation of protein samples: Proteins from human acute T-cell leukemia cells (Jurkat) were extracted by non-denaturing lysis. 10 8 Add 4 mL of ice-bathed non-denaturing lysate to each Jurkat cell, and the composition of the lysate is as follows: 50 mM 4-hydroxyethylpiperazineethanesulfonic acid (pH=7.4), 1 mM dithiothreitol, 150 mM sodium chloride, 1% ( v / v) Nonionic surfactant Triton X-100, 1mM ethylenediaminetetraacetic acid, 2% (v / v) protease inhibitor cocktail (Sigma), 2 / 5 tablets of phosphatase inhibitor (PhosSTOP , Roche), 1 mM phenylmethylsulfonyl fluoride. The ultrasonic power in the ice bath was 400W, the ultrasound was performed for 3s, the interval was 5s, and the ultrasound was repeated 180 times. Then centrifuge at 16000g centrifuge for 20min at 4°C, take the supernatant protein, measure the protein concentration to 6.5mg / mL according to the BCA protein co...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap