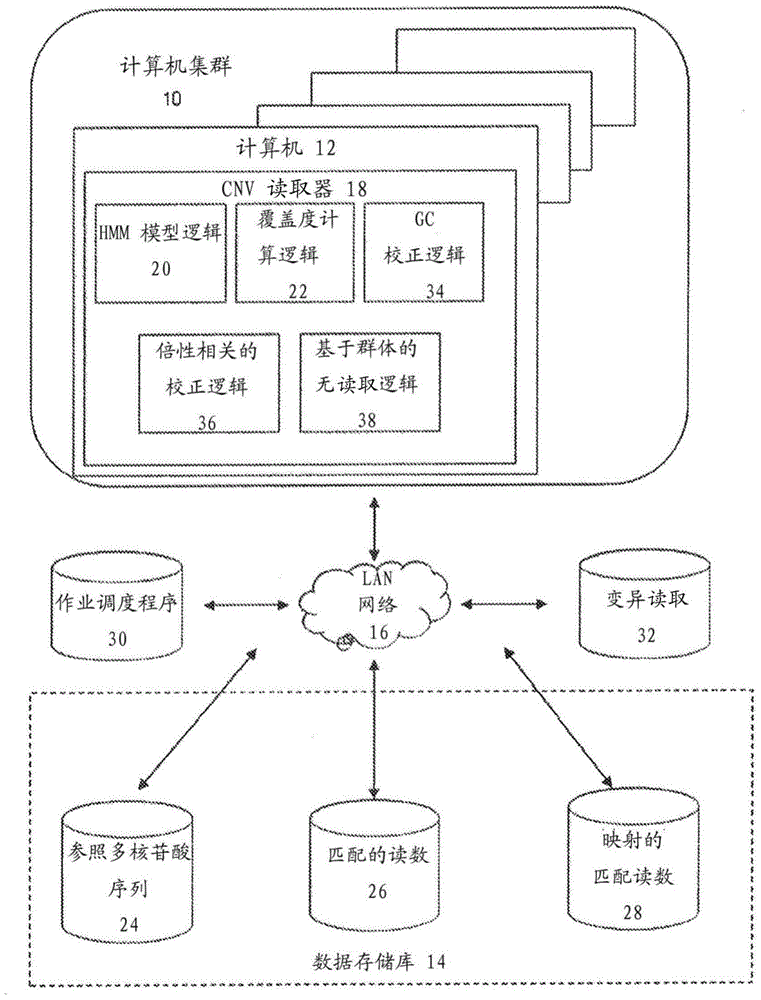
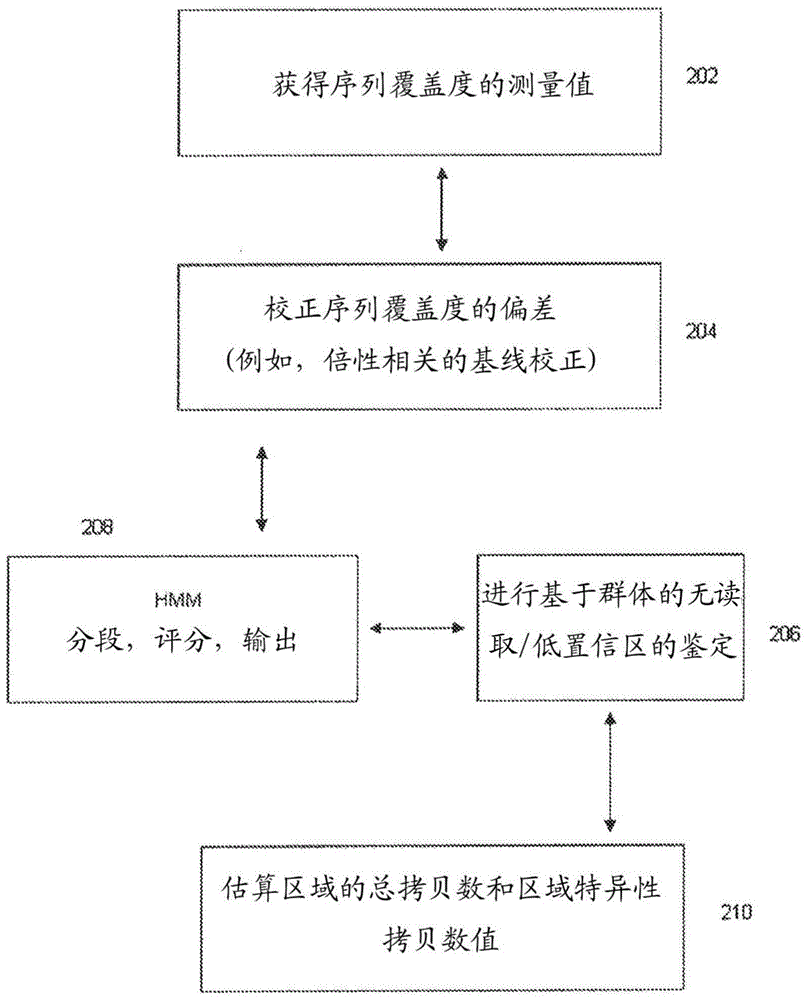
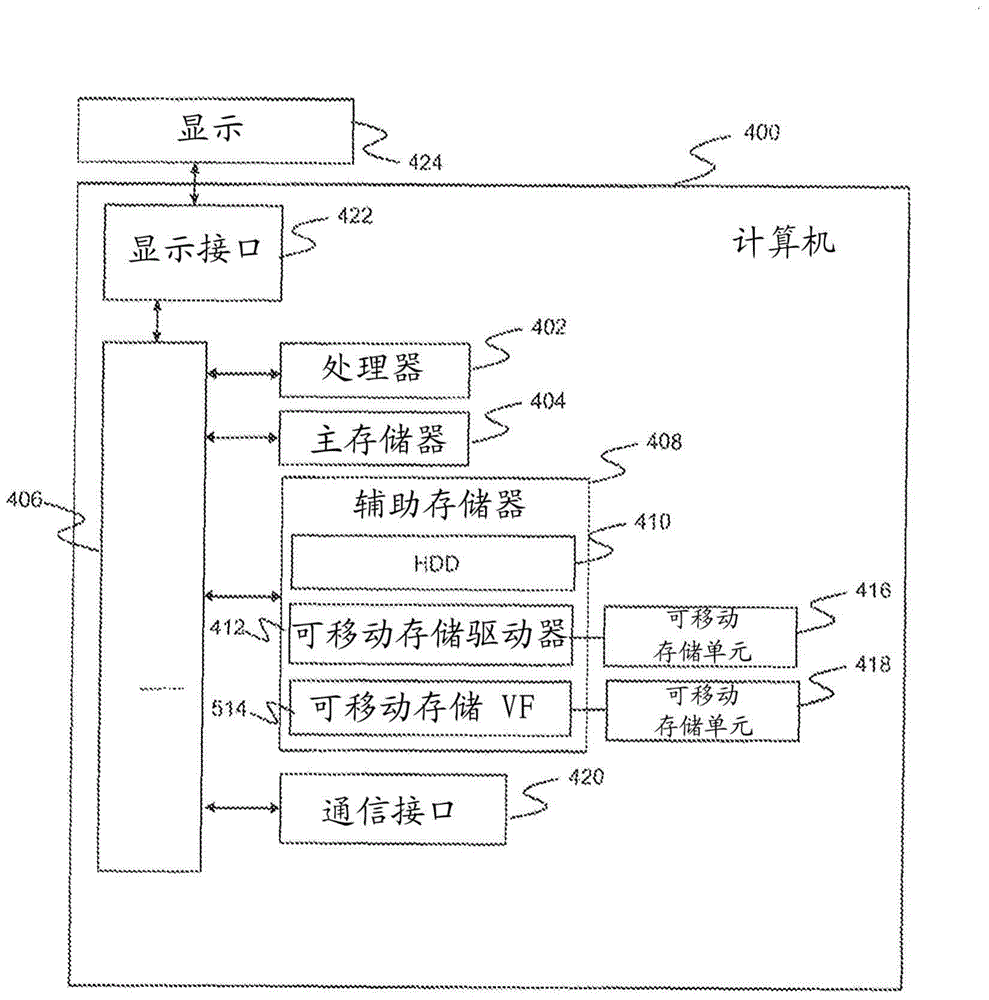
Methods for determining absolute genome-wide copy number variations of complex tumors
A genome and copy number technology, which is applied in the fields of genomics, biochemical equipment and methods, and the determination/inspection of microorganisms, which can solve the problems of error-prone data and human bias.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0041] In the following description, numerous specific details are set forth in order to provide a more thorough understanding of the present invention. It will be apparent, however, to one skilled in the art that the present invention may be practiced without one or more of these specific details. In other instances, features and procedures that are known to those skilled in the art have not been described in order to avoid obscuring the present invention.
[0042] Although the invention has been described primarily with reference to specific embodiments, it is contemplated that other embodiments will become apparent to those skilled in the art after reading this disclosure, and it is intended that such embodiments be included within In the method of the present invention.
[0043] Unless defined otherwise, all technical and scientific terms used herein have the same meaning as commonly understood by one of ordinary skill in the art to which this invention belongs. All publ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com