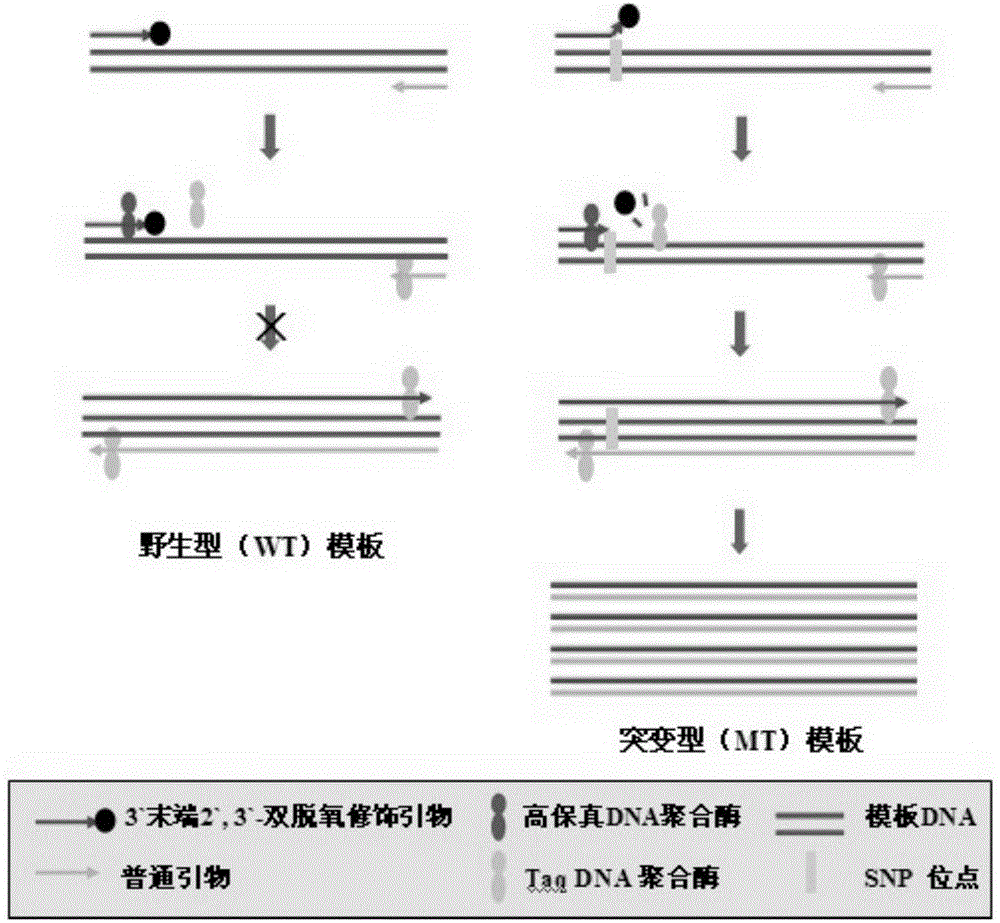
Rapid sensitive breast cancer susceptibility gene SNP detection method based on PCR primer 3' terminal nucleotide dideoxy modification
A susceptibility gene and detection method technology, applied in the field of rapid and sensitive detection of breast cancer susceptibility gene SNP
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
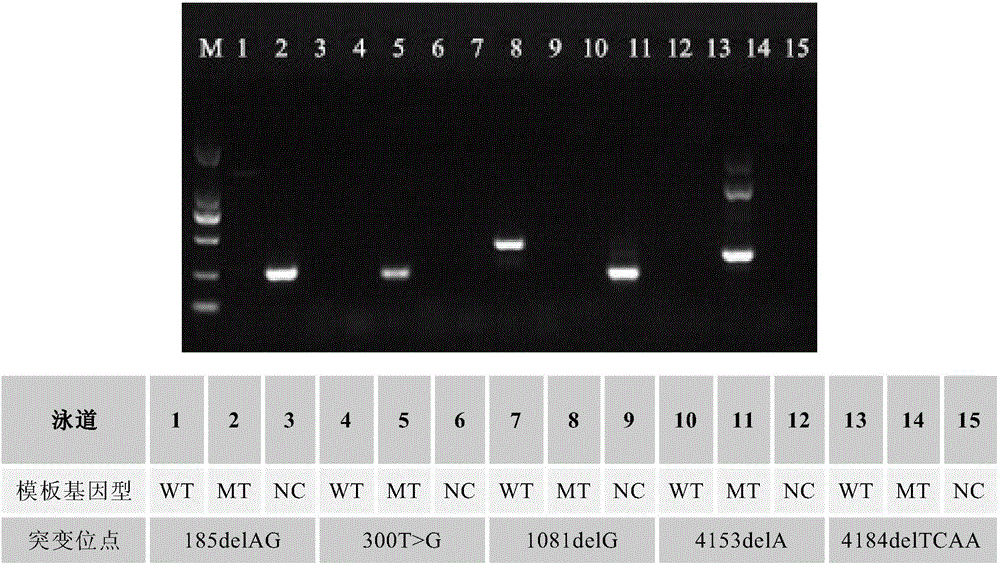
[0085] Example 1. Detection of hotspot base mutations in BRCA1 gene by the SNP detection method of breast cancer susceptibility genes based on PCR primer 3' terminal nucleotide dideoxy modification
[0086] In the following PCR detection methods, the template sequences of WT and MT used are the sequences of SEQ ID NO: 116-124.
[0087] (1) Primer design
[0088]In this example, the 185delAG mutation on exon 2, the 300T>G mutation on exon 5, the 1081delG mutation, 4153delA mutation and 4184delTCAA mutation on exon 11 of the BRCA1 gene hotspot mutation are taken as examples to analyze the mutations described in the present invention. Methods (300T>G, 1081delG, 4153delA and 4184delTCAA, and the names of the SNP sites to be detected involved in the following examples follow the customary naming of the mutation sites in known literature).
[0089] First, select 5 pairs (actually 9) of sequence fragments based on the full-length sequence of the BRCA1 gene, which are 185delAG wild-t...
Embodiment 2
[0101] Example 2. Detection of BRCA2 gene hotspot base mutations by the breast cancer susceptibility gene SNP detection method based on PCR primer 3' terminal nucleotide dideoxy modification
[0102] In the following PCR detection methods, the template sequences of WT and MT used are the sequences of SEQ ID NO: 130-135.
[0103] (1) Primer design
[0104] In this example, 999del5 on exon 9, 6174delT on exon 11 and 9254del5 on exon 23 of BRCA2 gene hotspot mutations are taken as examples to analyze the method of the present invention. First, 3 pairs of sequence fragments were selected according to the full-length sequence of the BRCA2 gene, which were 999del5 wild-type sequence, 999del5 mutant sequence, 6174delT wild-type sequence, 6174delT mutant sequence, 9254del5 wild-type sequence and 9254del5 mutant sequence (SEQ ID NO: 130~135). These three SNP sites are respectively covered, and compared with the wild-type sequence and the mutant sequence, there are only 1 to 5 base di...
Embodiment 3
[0114] Example 3. Sensitivity analysis of breast cancer susceptibility gene SNP detection method based on PCR primer 3' terminal nucleotide dideoxy modification
[0115] Taking the detection of the mutation 6174delT as an example, the wild-type (WT) sequence template (SEQ ID NO: 132) and the mutant (MT) sequence template (SEQ ID NO :133), the two DNA sequence fragments were connected to 18-T Vector (Dalian TaKaRa Company), then transformed the recombinant plasmid into DH5α competent cells, picked a single colony to expand culture, and screened out positive clones by bacterial liquid PCR, and sent them to Shanghai Boshang Biotechnology Co., Ltd. for sequencing. Finally, recombinant plasmids with completely correct sequences were selected. The two recombinant plasmids were digested with restriction endonuclease SacI (Dalian TaKaRa Company), respectively, and the linearized plasmids obtained after digestion were purified, and then the OD values were measured with an ultra-mic...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com