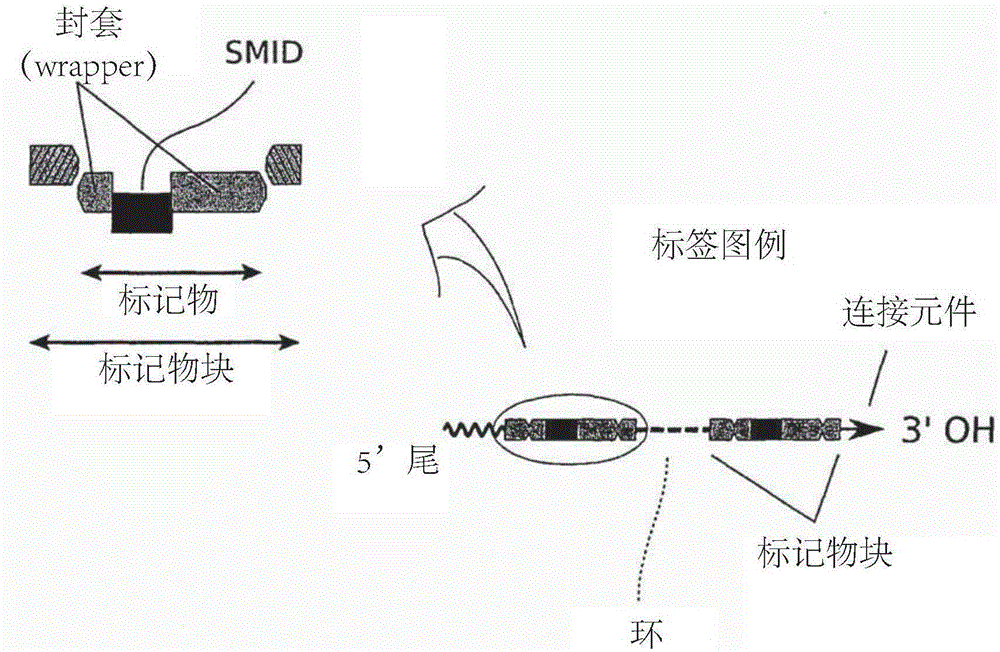
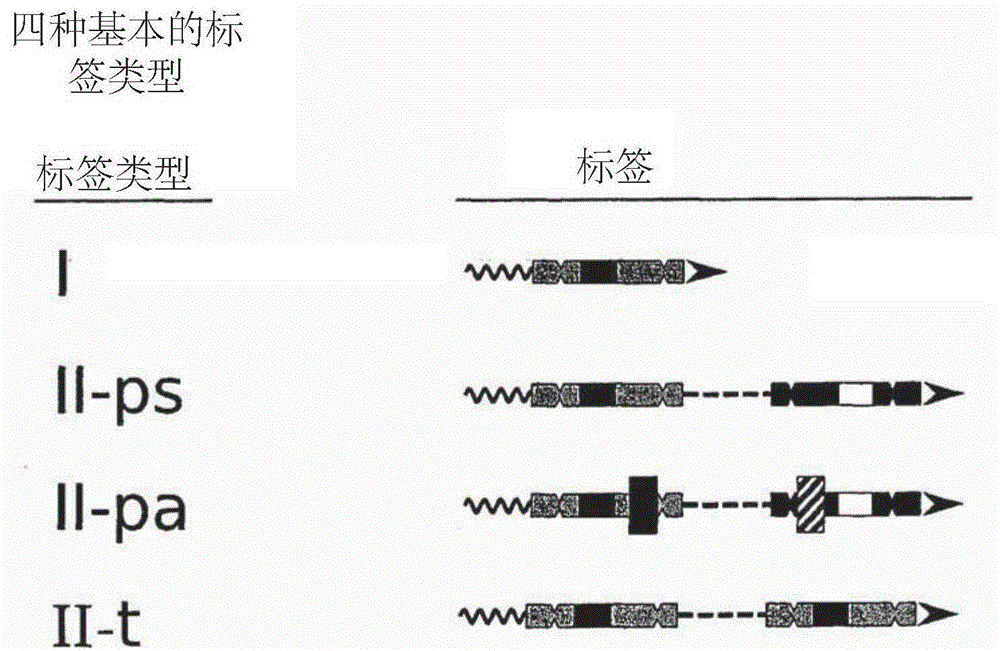
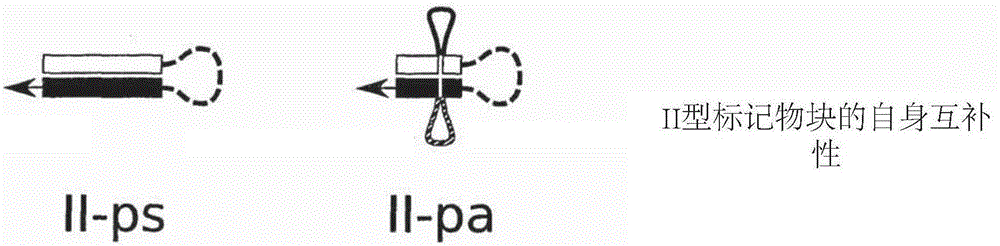
Methods of sequencing nucleic acids in mixtures and compositions related thereto
A technology of mixtures and compositions, applied in biochemical equipment and methods, measurement/testing of microorganisms, recombinant DNA technology, etc., can solve the problems of indistinguishable transcripts and incomplete sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0316] Example 1: Sequencing mRNA using labeling reagents
[0317] Isolation of poly-AmRNA from cells and tissues using standard kits and removal of residues of genomic DNA is routine (DNA-Free™, Life Technology).
[0318] 1. Reverse transcribe cDNA from RNA (Moloney Murine Leukemia Virus Reverse Transcriptase), prime with a labeling reagent containing SMID; treat the heteroduplex with RNase H. Moloney murine leukemia virus reverse transcriptase may be replaced by other viral reverse transcriptases or equivalent enzymes from any other source capable of reverse transcription of RNA.
[0319] 2. Circularize labeled single-stranded cDNA (T4RNA, DNA ligase (CircLigase; Epicentre)); use Exonuclease I to remove remaining linear cDNA.
[0320] 3. Divide the circularized cDNA suspension into aliquots and subject them to rolling circle amplification (RCA) (phi29 DNA polymerase) [The population of cDNA to be amplified may vary with the choice of primers. ]
[0321] 4. Optionally debr...
Embodiment 2
[0323] Example 2: Non-polyadenylated RNA
[0324] Salzman, J. et al reported in PloSOne, 2012, vol7, issue2, e30733 that circular RNAs are major transcript isoforms of hundreds of human genes in various cell types. These RNAs are not polyadenylated. Such RNA products are suitable for sequencing by this technique using labeling reagents of low stoichiometry with random 3' end sequences to obtain copies of the RNA, which are then circularized and processed as described herein.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com