Method for constructing fingerprint of phaseolus vulgaris according to SSR (simple sequence repeat) molecular marker and application
A technology of fingerprints and molecular markers, applied in biochemical equipment and methods, microbiological measurement/inspection, DNA preparation, etc., can solve the problems of distinguishing different bean varieties, different classification and naming standards of bean varieties, and achieve Short time consumption, accurate and reliable identification results, and effects that are not affected by the environment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] In this example, 36 kidney bean varieties were used as sample materials, and the fingerprint data of kidney bean varieties were constructed using the screened SSR core primers, so as to achieve the purpose of variety identification.
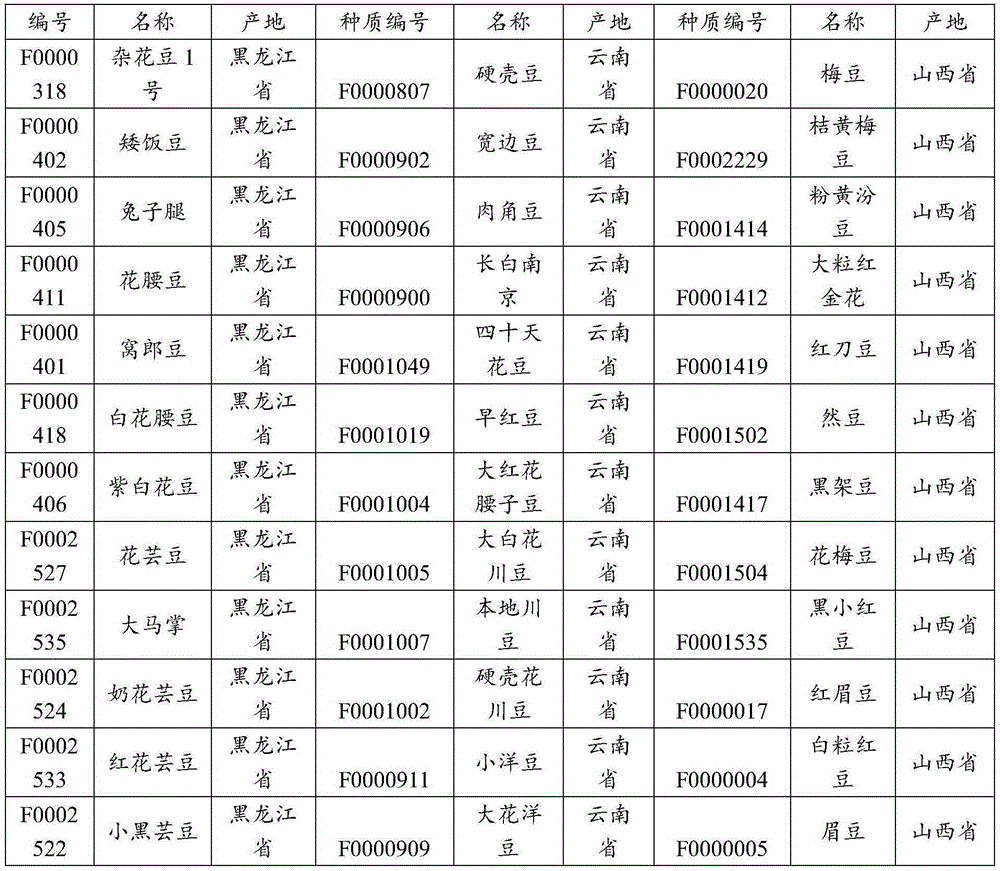
[0032] The information on the 36 tested kidney bean varieties is shown in Table 1 below. The germplasm numbers in Table 1 are from the China Crop Germplasm Information Network.
[0033] Table 136 information on the tested kidney bean varieties
[0034]
[0035] The construction of the standard DNA fingerprint library of kidney bean varieties is carried out according to the following steps:
[0036] 1. Extraction of total DNA from each variety of kidney bean
[0037] The bean was planted in the soil and cultured at room temperature. After 12 days of growth, the young bean leaves were taken and stored at -80°C after being frozen in liquid nitrogen. Take 0.2 g of tender young leaves of various varieties of kidney beans, put them in a 1....
Embodiment 2
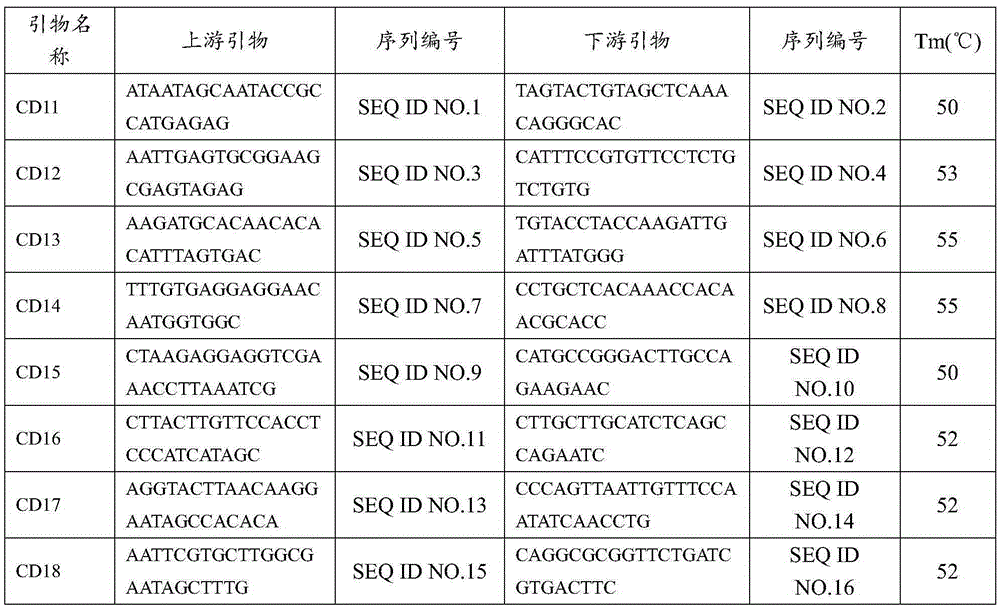
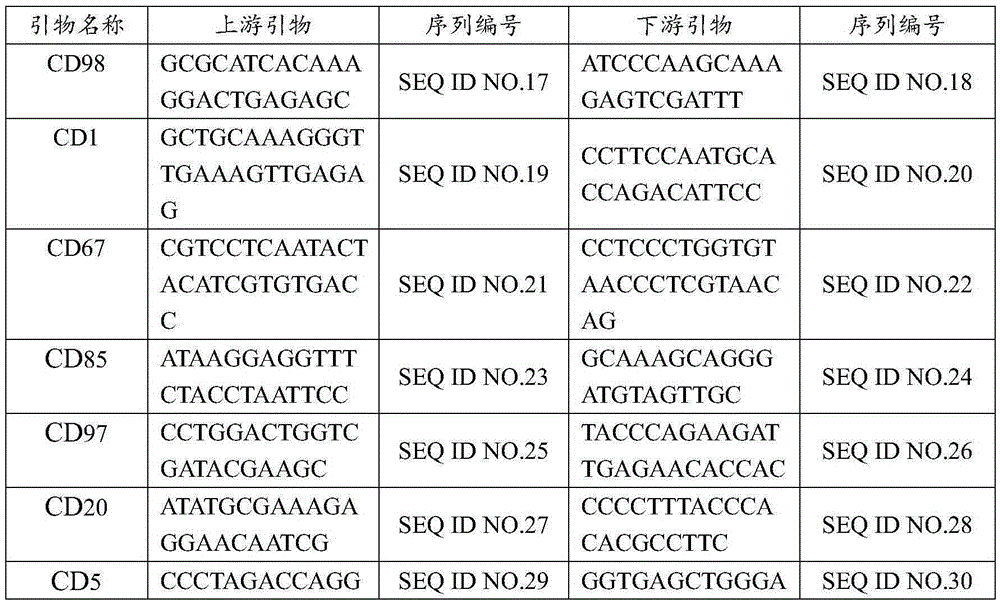
[0060] Example 2 Primer polymorphism verification
[0061] 40 pairs of primers except the core primers were used to amplify 36 samples of kidney bean by PCR to construct the fingerprint of kidney bean. Statistical primer polymorphism information, the results of which 8 pairs of non-core primers are listed below are the fingerprints of 6 varieties. The information of 8 pairs of non-core primers is shown in Table 6, and the fingerprint codes of 5 tested varieties are shown in Table 7. Shown, fingerprint code primer sequence CD98, CD1, CD67, CD85, CD97, CD20, CD5, CG1
[0062] Table 5 Partial non-core primer sequences
[0063]
[0064]
[0065] Table 6 Non-core primer information
[0066] Primer name number of alleles PIC CD98 1 0 CD1 2 0.0912 CD67 2 0.121 CD85 1 0 CD97 1 0 CD20 1 0 CD5 1 0 CG1 2 0.151
[0067] Table 75 Fingerprint Codes of Varieties
[0068] breed name fingerprint co...
Embodiment 3
[0071] Take 10 kidney bean varieties (different from the place of origin in Example 1) as verification materials as shown in Table 8, use the core primers screened to amplify 10 kidney beans, and use the method in Example 1 to construct fingerprints for anonymous samples. The code is shown in Table 8.
[0072] Table 810 Verified Bean Varieties
[0073]
[0074]
[0075] Table 910 Species Fingerprint Codes
[0076] Numbering breed name fingerprint code 1 yellow kidney beans 4-3-3-2-2-2-3-1 2 Noodles 2-1-4-1-3-5-2-3 3 flower kidney beans 2-3-4-2-3-2-2-1 4 flower kidney beans 1-4-1-2-3-2-3-1 5 plum beans 1-4-3-2-2-4-3-3 6 big horseshoe 2-2-2-4-2-5-2-2 7 hard shell beans 2-3-1-1-2-4-1-1 8 Flower face beans 1-2-5-4-3-4-2-4 9 Milky Kidney Beans 1-1-2-3-1-5-1-4 10 Flower face beans 1-4-4-3-1-1-1-1
[0077] It can be seen from Table 8 and Table 3 that plum beans and eyebrow beans in Example 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com