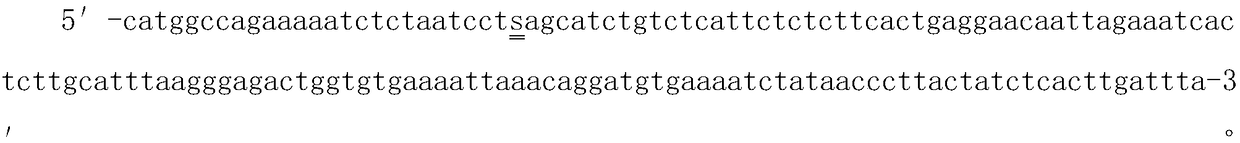A SNP molecular marker associated with growth traits of Pelteobagrus vachelli and application thereof
A technique for the growth traits of peltachlox vacheronii, applied in the field of aquatic animal genetics and molecular marker-assisted selection breeding, can solve the problems of wild resources and germplasm decline, seedling quality varies, and head length/body length increase, etc. Achieve accurate and reliable identification results, promote healthy development, and promote the effect of breeding progress
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] The SNP molecular marker of the present invention is derived from the genetic map of the F1 progeny constructed by our laboratory of Peeled catfish, and then obtained through the QTL mapping of growth traits (body weight, body width, and full length):
[0027] (1) Mining SNPs molecular markers and constructing a high-density genetic linkage map, using parents from different geographical groups and segregated traits, establishing the F1 generation family of Peltedelachus vacherii for > 3 months under the same conditions, and using microsatellite markers to screen A family with a high degree of heterozygosity was selected, and 200 full-sibling F1 generations and their parents were randomly selected as a mapping group, and the growth traits (weight, body width, total length, etc.) The -seq technology mines SNPs molecular markers and constructs a high-density genetic linkage map (Joinmap) of Pediatric catfish.
[0028] (2) QTL mapping for growth traits (body weight, body wi...
Embodiment 2
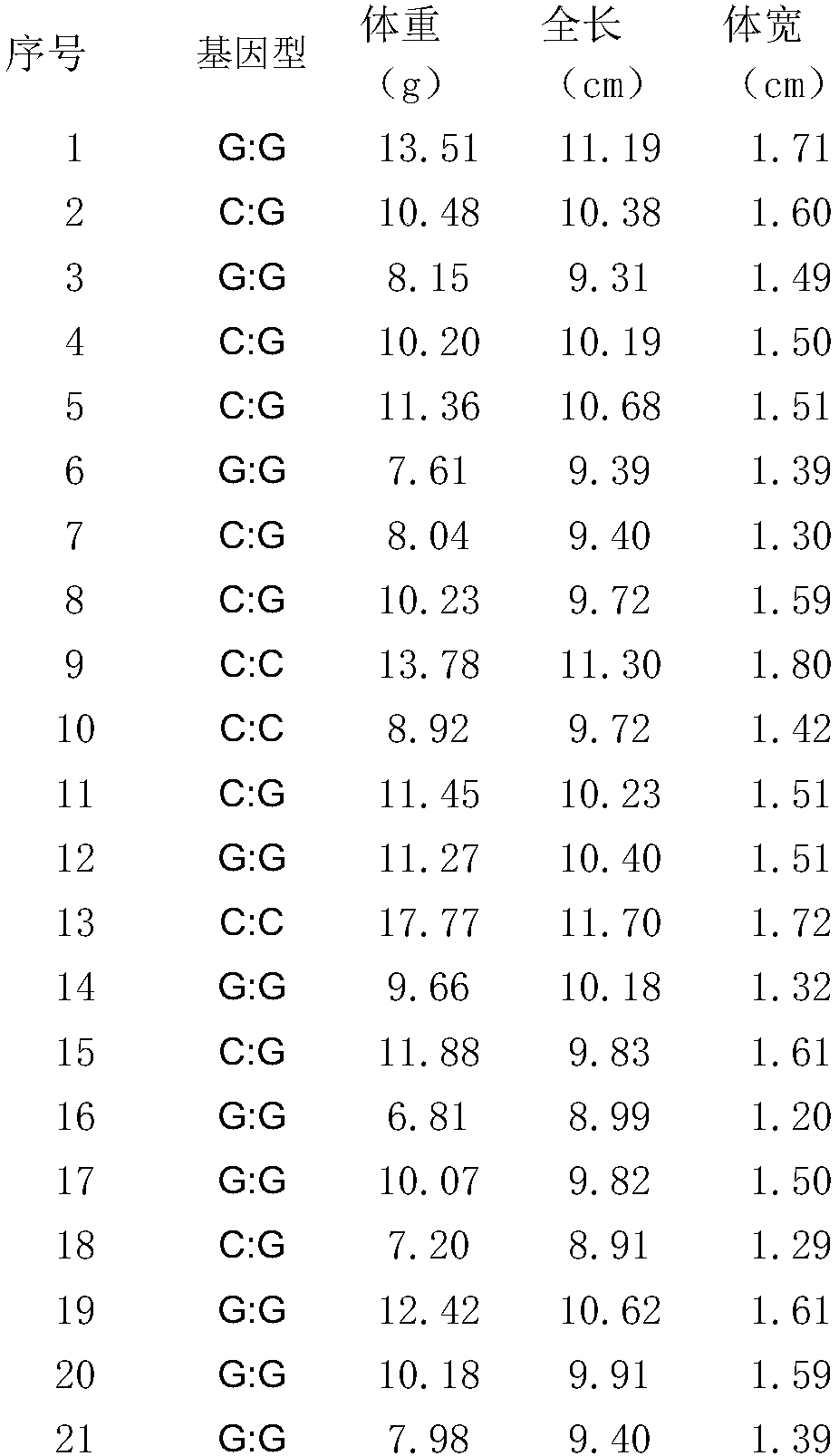
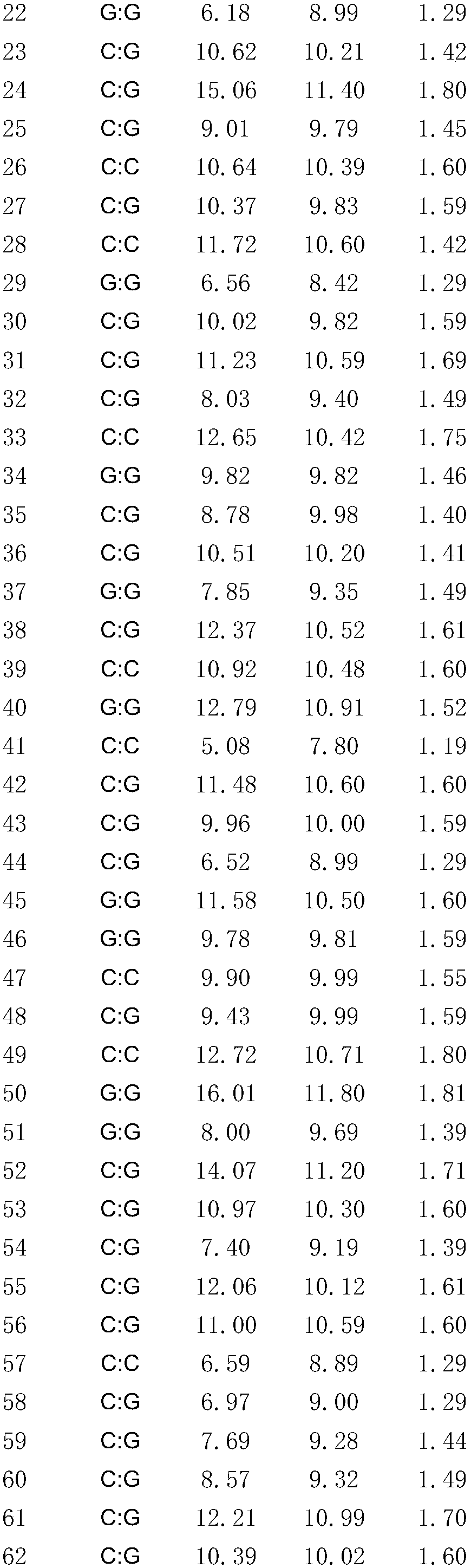
[0029] Example 2 Detection of the SNP molecular marker of the sample
[0030]Since the film was released in late July, 2017, 450 individual pelturotus vacheronii were cultured for 105 days under the same environment (temperature, density, etc.); the 450 fish were placed in a pool (580L) to adapt for 2 weeks, and carried out Stop feeding the feed for 2 days before the experiment, use a caliper to measure its body width and total length, and use an electronic balance to measure its body weight; randomly select 193 Pelvis catfish, cut their tail fins and store them in 95% ethanol at -20°C for use in Genomic DNA extraction, the specific steps are as follows:
[0031] (1) Take a small amount of caudal fins and extract the genomic DNA of Peeled catfish;
[0032] (2) PCR amplification is carried out by using a specific primer set, and the resulting amplification product is scanned for fluorescent signals;
[0033] (3) Determine whether the genotype of the SNP site at the 26bp posit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com