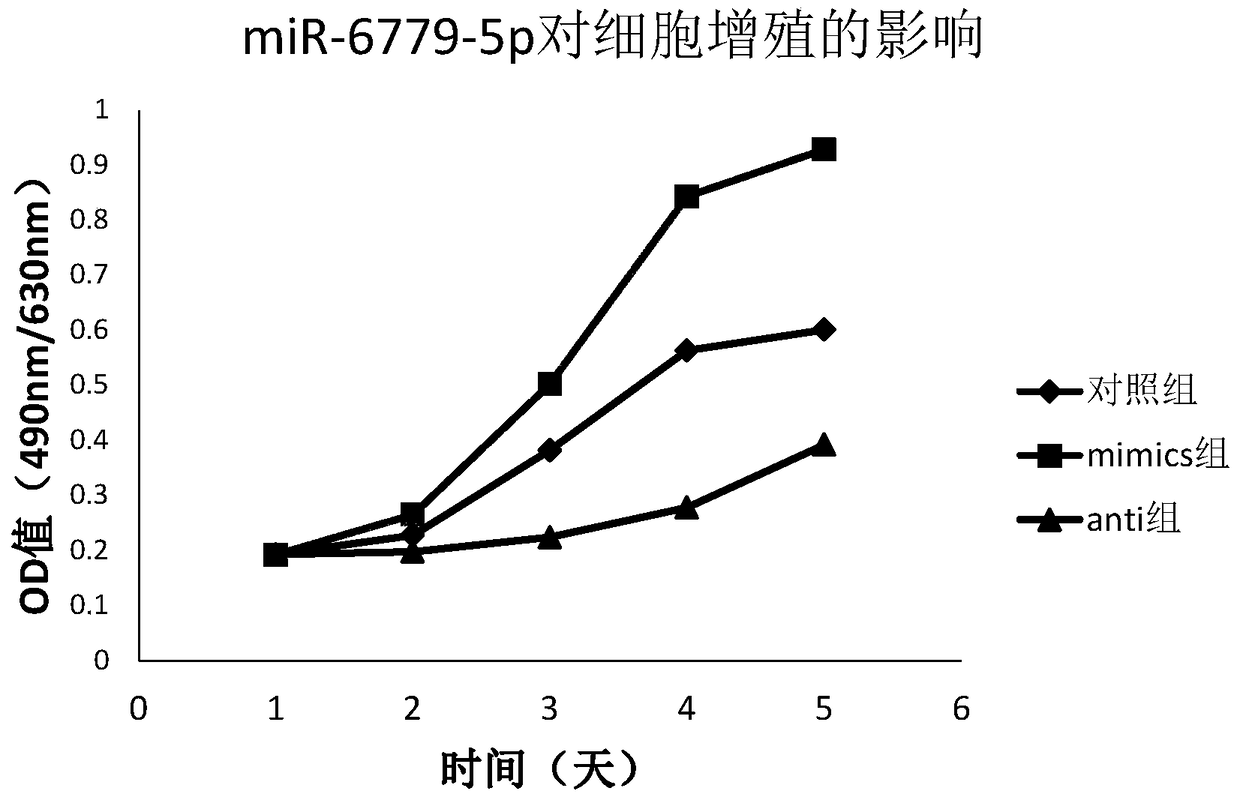
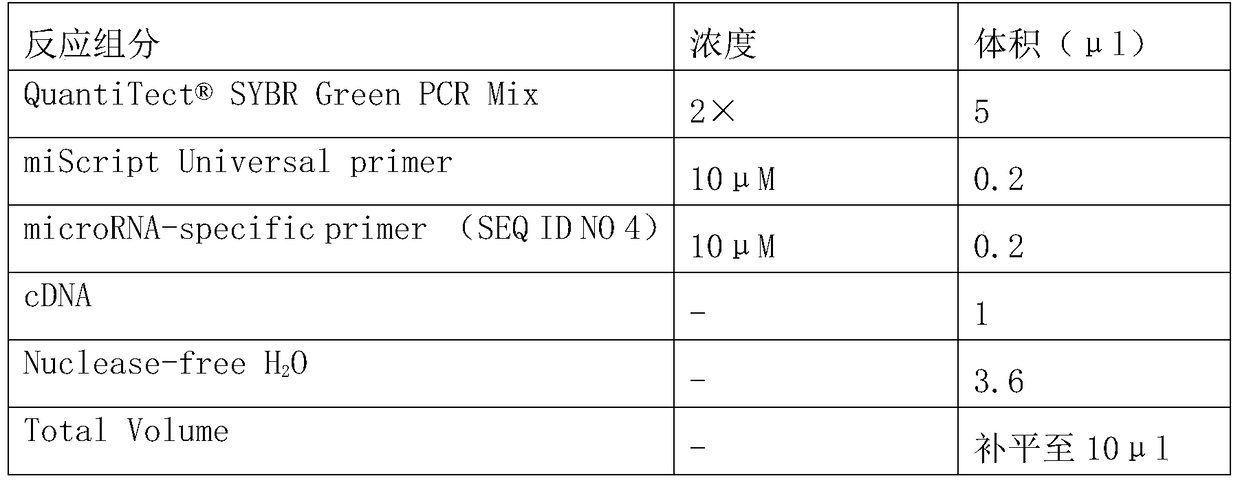
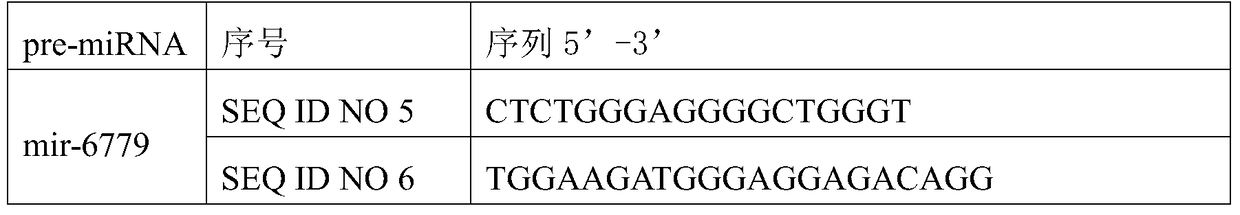
A preparation for diagnosis and treatment of osteosarcoma and its application
An osteosarcoma and preparation technology, applied in the field of molecular biology, can solve the problem of stagnant 5-year survival rate of osteosarcoma
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] The collection of embodiment 1 sample
[0057] 7 patients with primary osteosarcoma and 8 healthy controls. The patients with primary osteosarcoma and the healthy control group were required to fast for at least 12 hours. At 7:00-8:00 the next morning at room temperature, 10ml of venous blood was drawn into ethylenediaminetetraacetic acid (EDTA) anticoagulant tubes, and peripheral blood was extracted. For mononuclear PBMCs, add 1ml Trizol reagent (Invitrogen), mix thoroughly, and store the specimen at -80°C for RNA extraction. All blood samples and pathological results should be authentic and reliable, the study was approved by the ethics committee, and the patients gave informed consent.
Embodiment 2
[0058] Example 2 total RNA extraction
[0059] Total RNA was extracted from peripheral blood mononuclear cells (PBMCs) of patients and healthy controls.
[0060] Requirements: RNA purity: OD260 / 280≧1.8, 28S / 18S≧1; RNA integrity: RIN value≧7.0.
[0061] Method: RNA concentration and purity detection method: NanoDrop2000;
[0062] RNA integrity detection method: Agilent 2100 (RNA 6000Nano kit), agarose gel electrophoresis (agarose gel concentration: 1% agarose gel; voltage: 5V / cm; time: 20min).
[0063] Small RNA sequencing sample requirements: concentration ≥ 200ng / μL, total amount ≥ 10μg of RNA (5 μg for a single library construction); OD260 / 280 between 1.8 and 2.2, OD260 / 230 ≥ 2.0, RIN ≥ 6.5, 28S :18S≥1.0, no RNA degradation.
Embodiment 3
[0064] Example 3 Sequencing and Data Analysis
[0065] The sequencing company carried out the establishment of the sequencing library and on-machine sequencing, and the sequencer used was the HiSeq2000 sequencer of Illumina Company.
[0066] The miRNA raw data were background-corrected by transcriptome data analysis software, and then the t-test was performed to obtain the P value, and then the Fisher test was used to combine the P values to screen for differentially expressed miRNAs. Set p value2, and then artificially select and filter differentially expressed miRNAs up-regulated miR-6779-5p (log2(Fold_change) value is 2.16) into our research scope.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com