Method for locating sequence position causing special growth phenotype of yeast
A yeast and phenotype technology, applied in the field of locating the sequence position that causes the special growth phenotype of yeast, can solve the problems of time-consuming and labor-intensive, and achieve the effect of avoiding time-consuming and labor-intensive
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1: Sequence mapping of semisynthetic yeast yYW0062
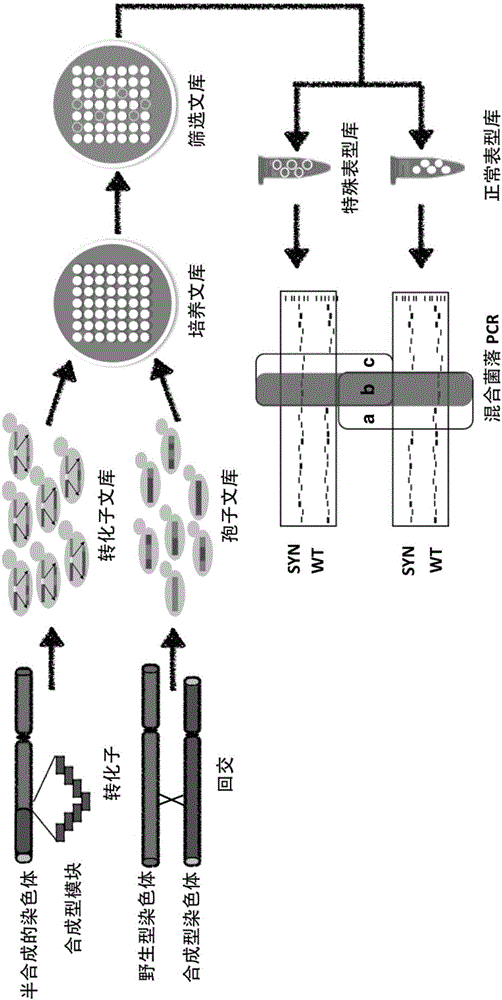
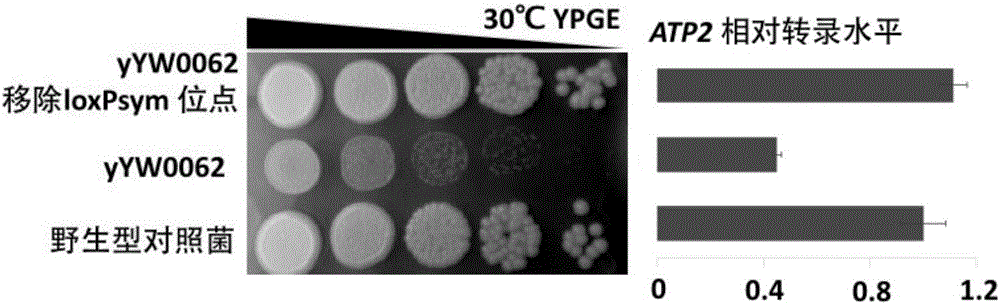
[0048] The semi-synthetic yeast strain yYW0062 (a semi-synthetic yeast strain obtained from wild-type S. BY4741 does not exist (see figure 2 ). The transformants produced during the integration of the synthetic module were collected to construct a transformant library, and the transformant library was copied onto the YPGE medium, and the growth of the transformants on the YPGE was observed after 2 days. Selective culture conditions were set according to the above-mentioned special growth phenotypes, and the transformants could be divided into two types under the condition of selection pressure: transformant pools that grew normally on YPGE, and transformant pools that had growth defects on YPGE. Use the PCR tags unique to the semi-synthetic yeast chromosome and the PCR tag specific to the wild-type yeast chromosome to design amplification primers for each functional gene of the two, and extract the mixed ge...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com