A computer simulation method to obtain the intermediate state structure of G protein-coupled receptors
A technology for coupling receptors and G proteins, which is applied in the field of computer simulation of G protein-coupled receptor structures, can solve problems such as limiting the structure and function of GPCRs, difficulty in obtaining all structures, and limited receptor structures, so as to save time and Effects of computer resources, resolution of simulation time constraints, simplicity of approach
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
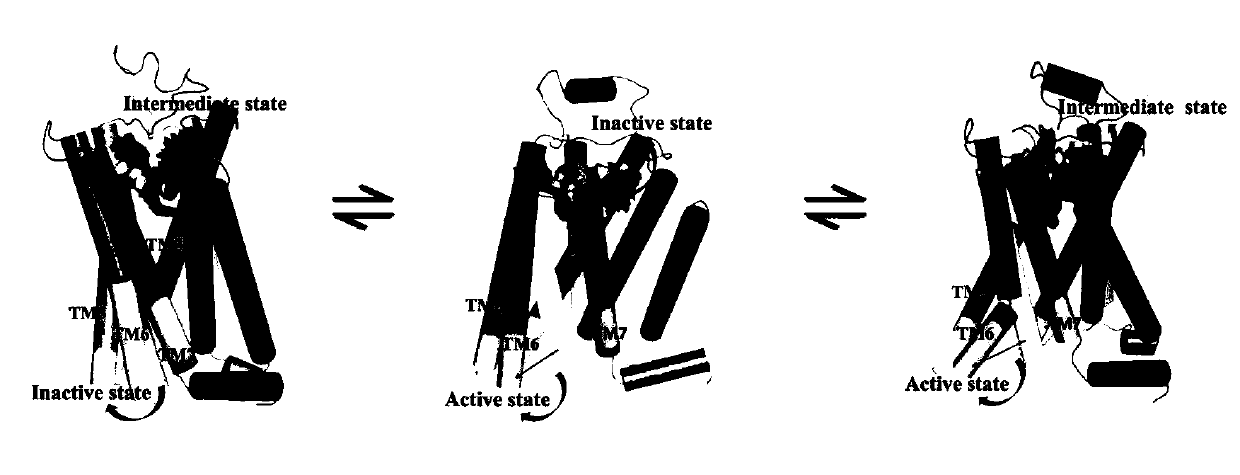
[0034] Embodiment 1 computer simulation obtains adrenoceptor (β 2 AR) intermediate state structure
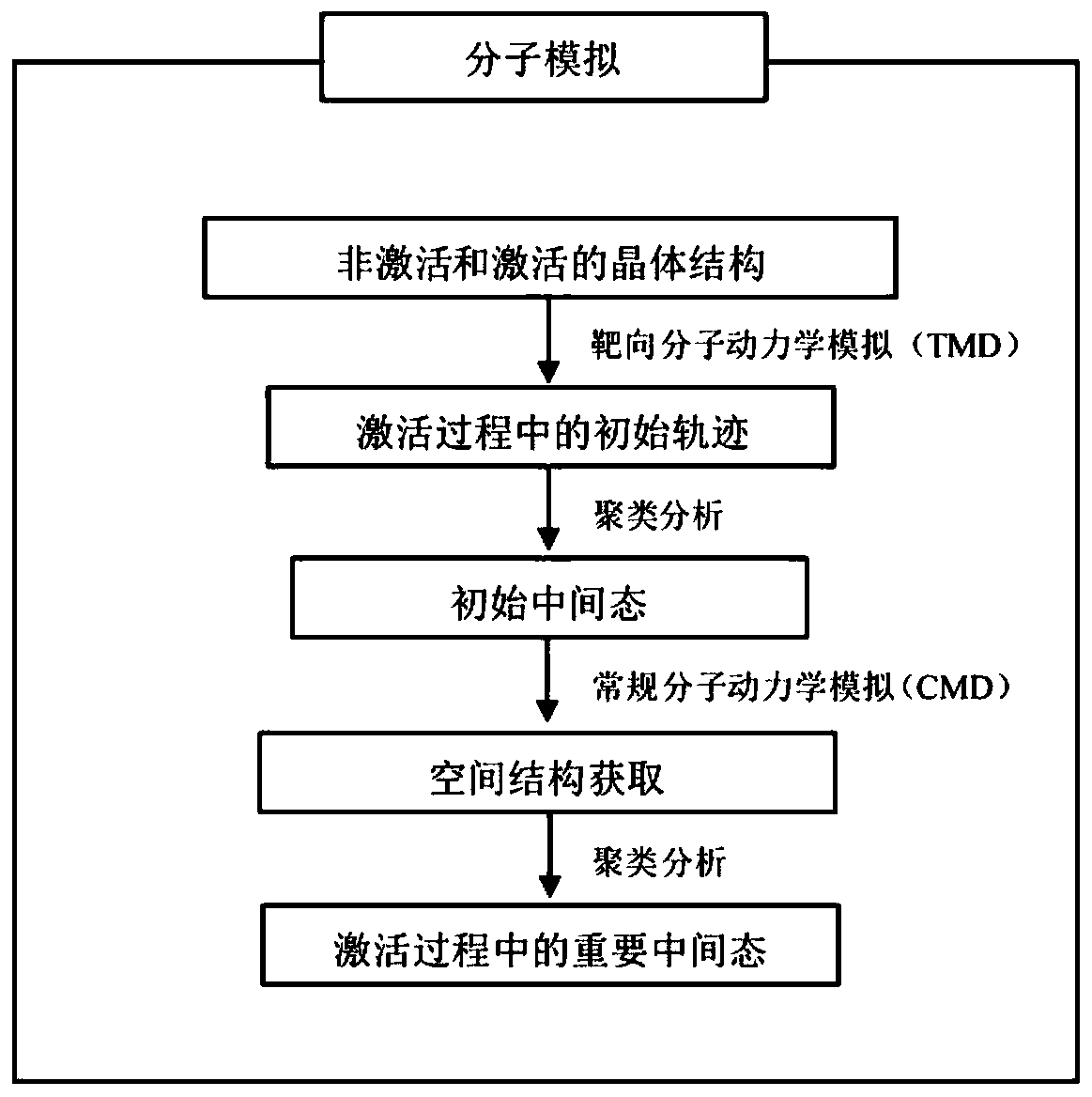
[0035] ①Download adrenergic receptor (β 2 AR) The crystal structures of the inactive and active states, PDBID numbers are 2RH1 and 3SN6, respectively.
[0036] ②In order to ensure that the number of atoms in the initial structure of the non-activated and activated structures is the same, remove all non-receptor molecules in the Pymol software, and finally obtain the ligand-free receptor (apo)
[0037] ③In CHARMM-GUI ( http: / / www.charmm-gui.org / ) into the phospholipid bilayer (POPC), water, ions, etc. to construct the simulated environment required for the receptor.
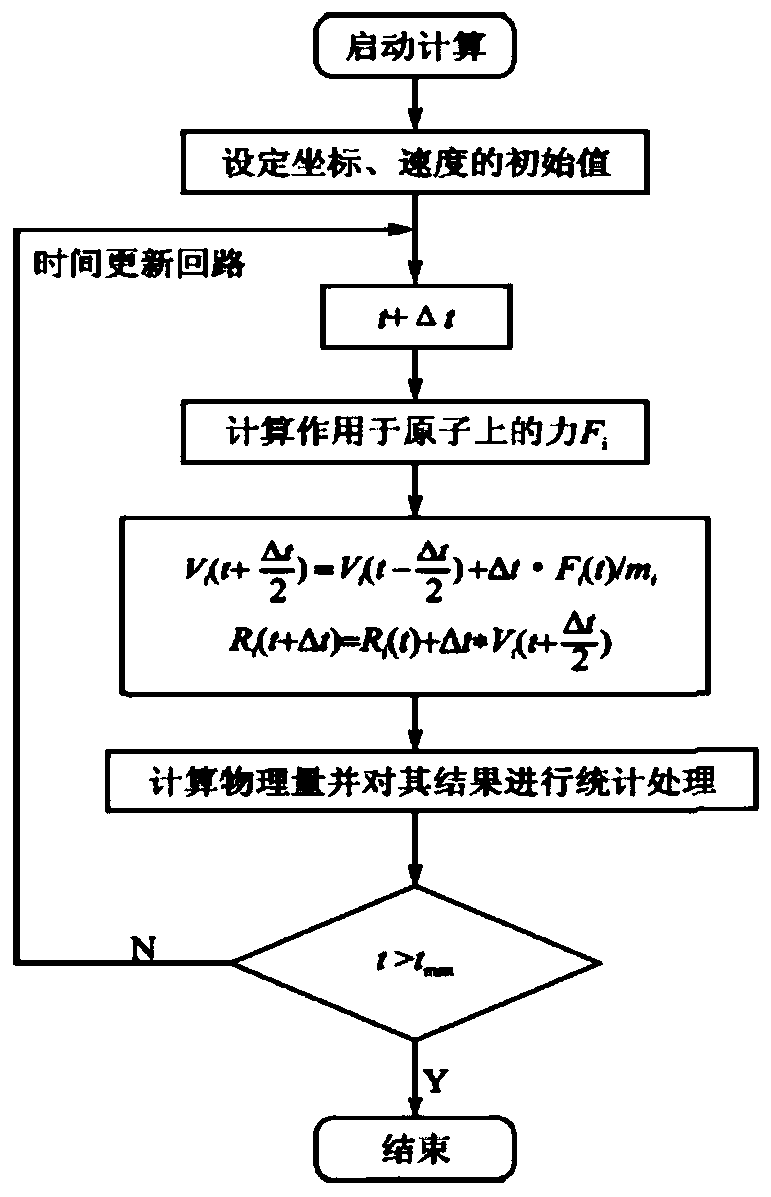
[0038] ④ There may be many unnatural contacts in the randomly constructed system, and the potential energy of the system will be very high. Therefore, in order to eliminate the unreasonable situation of the atomic positions in the simulated system, in the Sander module of Amber16, the steepest descent method i...
Embodiment 2
[0047] Example 2 Computer simulation to obtain the intermediate state structure of μ-opioid receptor (μ-Opioid)
[0048] ①Download the inactive and active crystal structures of μ-opioid receptor (μ-Opioid) from the PDB database, and the PDB ID numbers are 4DKL and 5C1M, respectively.
[0049] ②In order to ensure that the number of atoms in the initial structure of the non-activated and activated structures is the same, remove all non-receptor molecules in the Pymol software, and finally obtain the ligand-free receptor (apo)
[0050] ③In CHARMM-GUI ( http: / / www.charmm-gui.org / ) to add phospholipid bilayer (POPC), water, ions, etc. to construct the simulated environment required by the receptor.
[0051] ④ There may be many unnatural contacts in the randomly constructed system, and the potential energy of the system will be very high. Therefore, in order to eliminate the unreasonable situation of the atomic positions in the simulated system, in the Sander module of Amber16, t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com