SNP molecular marker ITS296 for detecting dog-derived Ceylon hookworms and ancylostoma caninum, primer and application thereof
A technique of hookworm of ceylon and molecular marker is applied in the field of hookworm disease diagnosis and detection, which can solve the problems of high cost, inability to detect a large number of samples at the same time, complicated operation, etc., and achieve the effect of completely consistent accuracy and completely consistent sequencing results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
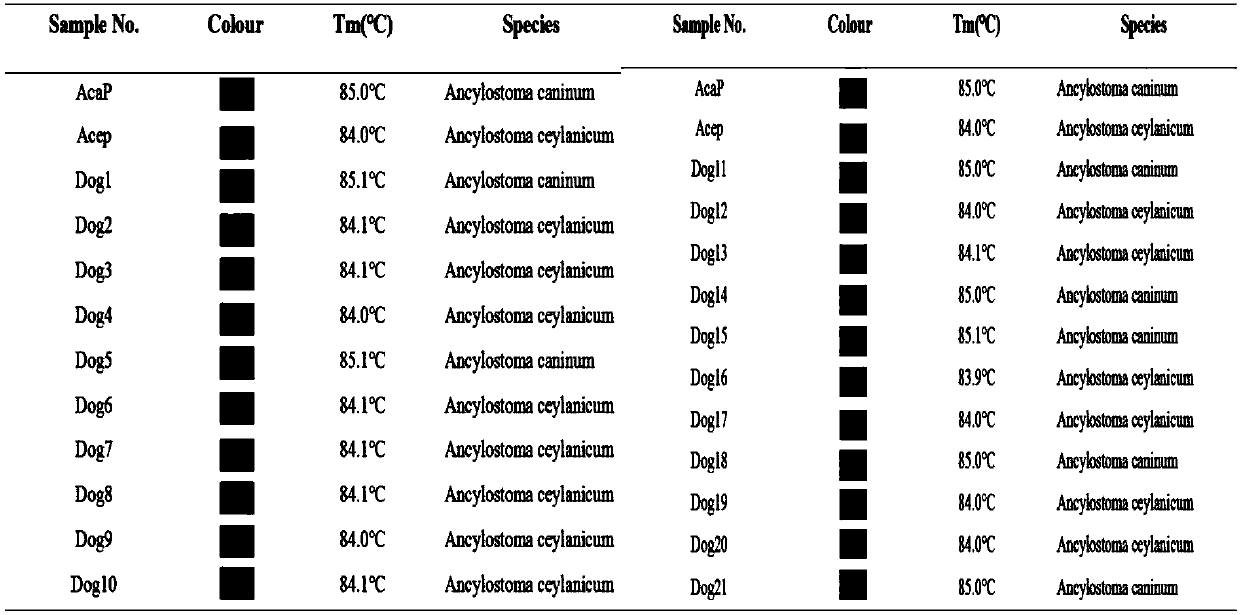
[0048] Example 1. Identification of Ancylostoma Ceylonium from dogs and Ancylostoma caninum
[0049] (1) First refer to the ITS sequences of Hookworm canine (LC177192.1) and Hookworm Ceylon (KM066110.1) downloaded from NCBI, and design a set of T based on a SNP site ITS296 of ITS1. m -shift PCR primers (2 forward specific primers and 1 common reverse primer), see Table 1.
[0050] Table 1 T based on ITS1 sequence SNP site ITS296 m -shift primer
[0051]
[0052] Establish T according to the above primers m -shift PCR reaction system (Table 2) and reaction conditions:
[0053] Table 2 T m -shift PCR reaction system
[0054]
[0055] Reaction conditions:
[0056]
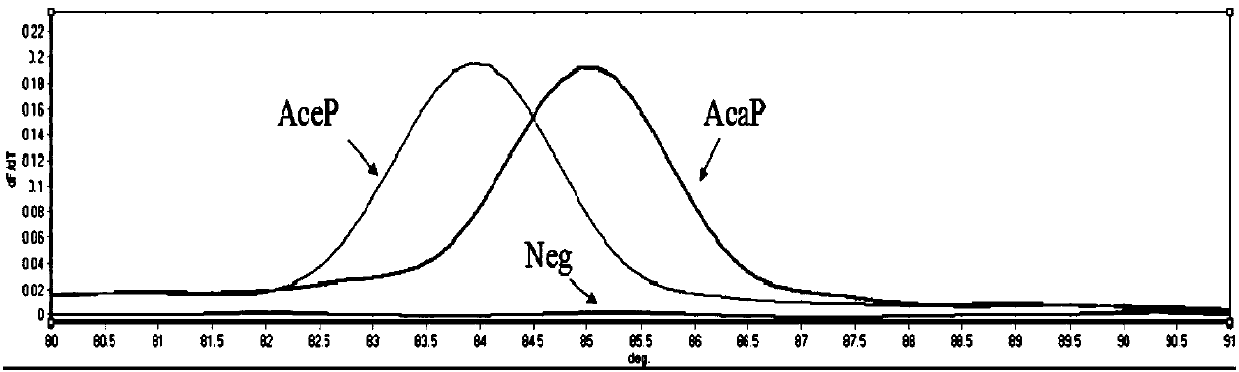
[0057] (2) T based on SNP site ITS296 m -shift primers for the standard plasmids of Hookworm Ceylon (AceP) and Hookworm (AcaP) m -shift standard curve see figure 1 . The results showed that the T based on SNP site ITS296 m The -shift method can distinguish Ceylon hookworm from dog hookworm. Through the analysis of Rotor-Gene Q...
Embodiment 2
[0058] Example 2.T m -shift method stability test
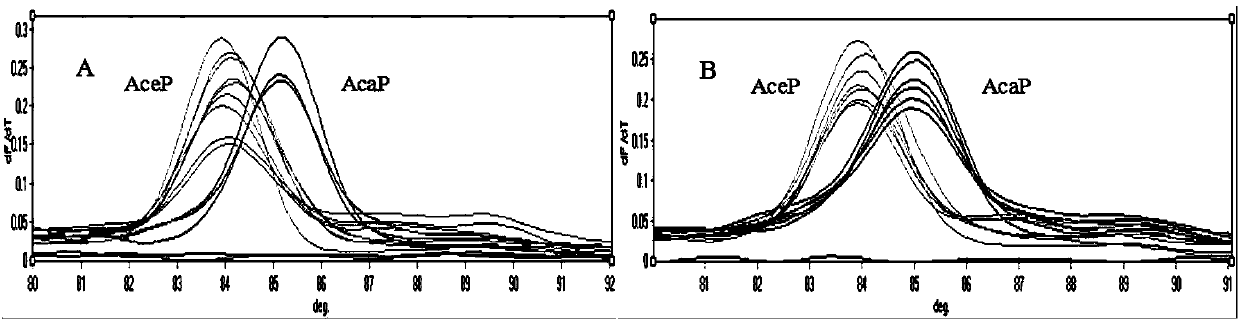
[0059] To evaluate the established T based on SNP locus ITS296 m -The stability of the shift method, the test carried out reproducibility testing on the constructed two known plasmid standards. Each sample is tested 7 times in batches, and 3 times between batches, qPCR-T m -shift reaction system and cycle parameters are the same as above. The test results showed that in the primers of ITS296, Ancylostoma canis and Ancylostoma T m The CV of the value is 0.07% and 0.13% respectively (Table 3), so the T m The -shift molecular detection method has good intra-batch and inter-batch reproducibility.
[0060] Table 3 Repeatability test results
[0061]
Embodiment 3
[0062] Example 3.T m -shift method sensitivity test
[0063] To detect established T based on SNP site ITS296 m -Sensitivity of the shift method. Dilute the prepared two plasmid standards by 10-fold ratio respectively, and press 1:10 to 1:10 8 Concentration is detected, qPCR-T m -shift reaction system and cycle parameters are the same as above. The results showed that the dilution of H. canis and H. Ceylon samples was as low as 1:10 7 (I.e. 5.33×10 -6 ng / μL and 5.03×10 -6 ng / μL), T m The -shift method can still distinguish the two (see Table 4).
[0064] Table 4 The T of different concentrations of Ancylostoma caninum and Ancylostoma Ceylonum m value
[0065]
[0066] Note: "-" means that the target peak cannot be detected
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com