SNP molecular marker ITS197 for detecting dog-derived Ceylon hookworms and ancylostoma caninum, primer and application thereof
An hookworm of ceylon and molecular marker technology, which is applied in the field of hookworm disease diagnosis and detection, can solve the problems of high cost, inability to detect a large number of samples at the same time, and complicated operation, and achieve the effect of completely consistent accuracy and completely consistent sequencing results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Embodiment 1. The identification of canine Ancylostoma ceyloni and Ancylostoma canis
[0049] (1) First, refer to the ITS sequences of Ancylostoma canis (LC177192.1) and Ancylostoma ceylonum (KM066110.1) downloaded from NCBI, and design a set of T m -shift PCR primers (2 forward specific primers and 1 common reverse primer), see Table 1.
[0050] Table 1 Based on the T of ITS1 sequence SNP site ITS197 m -shift primer
[0051]
[0052] According to the above primers to establish T m The reaction system (Table 2) and reaction conditions of -shift PCR:
[0053] Table 2 T m -shift PCR reaction system
[0054]
[0055] Reaction conditions:
[0056]
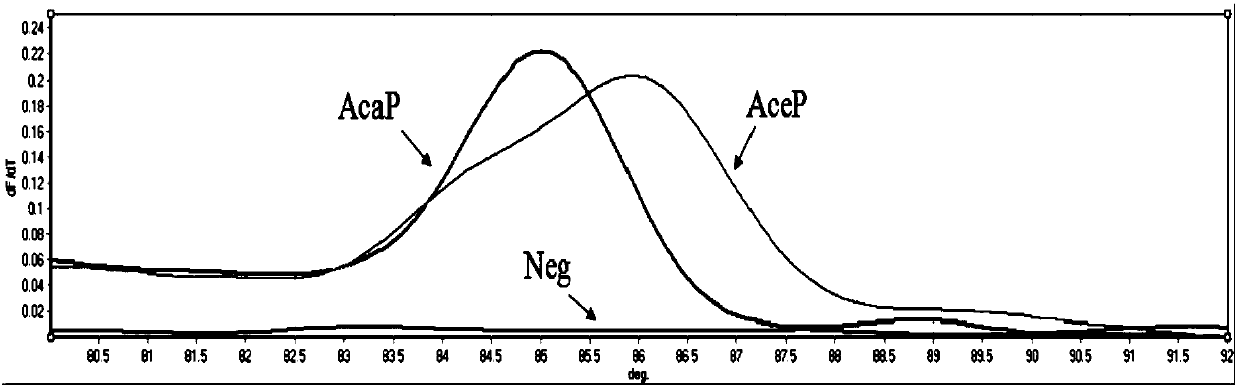
[0057] (2) T based on SNP site ITS197 m -shift primers for AceP and AcaP standard plasmids m -shift standard curve see figure 1 . The results showed that the T based on the SNP site ITS197 m The -shift method can distinguish Ancylostoma ceyloni from Ancylostoma canis. Through the analysis of Rotor-Gene Q ser...
Embodiment 2
[0058] Example 2.T m Stability test of -shift method
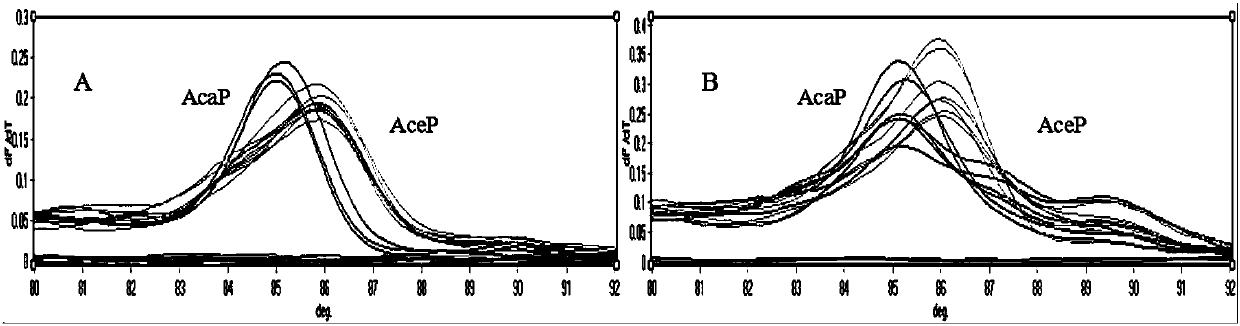
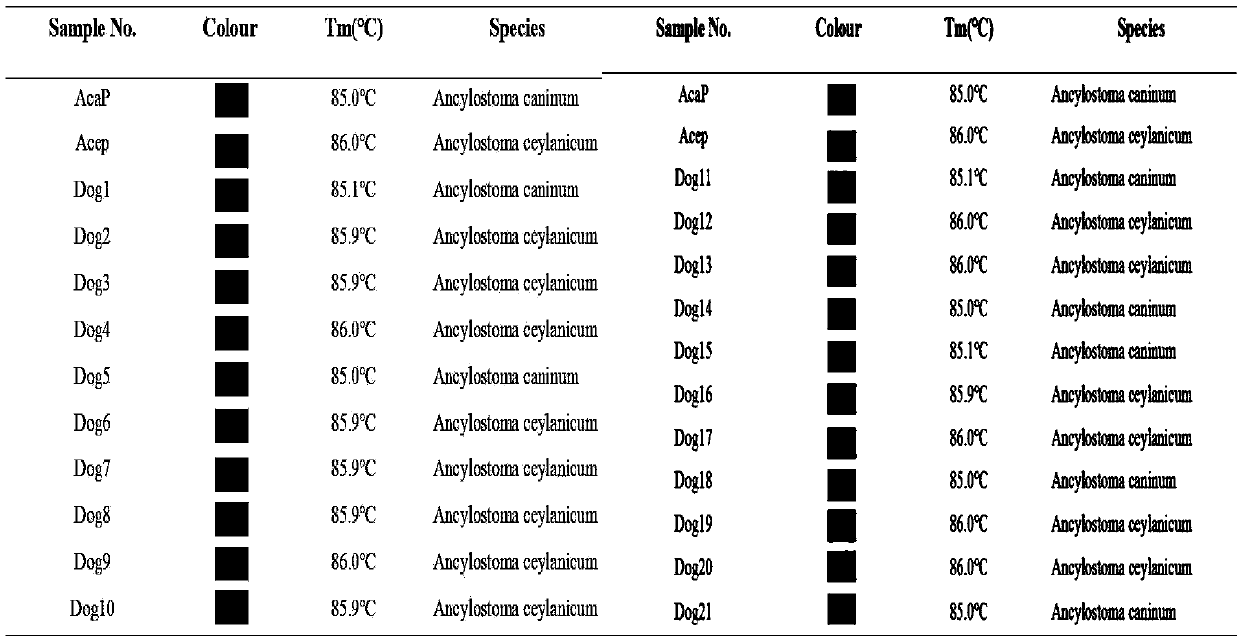
[0059] To evaluate the established SNP site ITS197-based T m The stability of the -shift method was tested for the repeatability of the two known plasmid standards constructed. Each sample was tested 7 times within a batch and 3 times between batches. qPCR-T m -shift reaction system and cycle parameters are the same as above. The test results showed that among the primers of ITS197, Ancylostoma caninum and Ancylostoma ceyloni T m The CVs of the values are 0.14% and 0.18% respectively (Table 3), so the T m The intra-assay repeatability and inter-assay repeatability of -shift molecular detection method were both good.
[0060] Table 3 Repeatability test results
[0061]
Embodiment 3
[0062] Example 3.T m Sensitivity test of -shift method
[0063] To detect the established SNP site ITS197-based T m The sensitivity of the -shift method is to dilute the two prepared plasmid standards by 10 times, according to the ratio of 1:10 to 1:10. 8 Concentration detection, qPCR-T m -shift reaction system and cycle parameters are the same as above. The results showed that when the sample dilution of Ancylostoma caninum and Ancylostoma ceylonis as low as 1:10 7 (i.e. 5.33×10 -6 ng / μL and 5.03×10 -6 ng / μL), T m The -shift method can still distinguish the two (see Table 4).
[0064] Table 4 primers to different concentrations of Ancylostoma caninum and Ancylostoma ceyloni T m value
[0065]
[0066] Note: "-" indicates that the target peak cannot be detected
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com