Plant RNA modification and editing system and method
A plant and editing technology, applied to the system and field of plant RNA modification and editing, to achieve the effect of an efficient and rapid RNA modification and editing system
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0175] 15. Preparation and transformation of Arabidopsis protoplasts
[0176] Take the young leaves of Arabidopsis thaliana, and use a blade to cut the leaves into 0.5mm-1mm size. Immerse in 10ml of enzymatic hydrolysis solution, mix well, and culture in dark for 2h-3h, until the protoplasts are completely dissociated from the leaves. After the enzymatic hydrolysis, check the enzymatic hydrolysis results under a microscope. Use a 200-mesh stainless steel mesh screen to filter the protoplasts into 10ml of a new centrifuge tube. Centrifuge at 100×g for 2 minutes at 4°C to collect protoplasts. Resuspend the protoplasts in an equal volume of pre-cooled W5 liquid medium, centrifuge at 100×g for 2 min, collect the protoplasts, and repeat 2 times. Resuspend the protoplasts in an equal volume of pre-cooled W5 liquid and keep on ice for 30 minutes. Centrifuge at 100×g for 2 minutes to collect the protoplasts and resuspend the protoplasts in 1 / 10 volume MMg. Take a small amount of ...
Embodiment 1
[0182] Example 1 In vitro methylation of dCas13a-RsmB
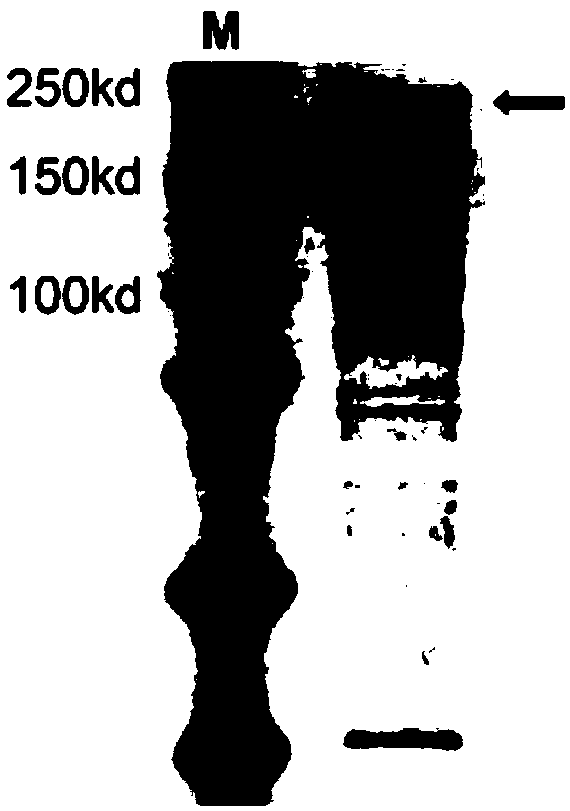
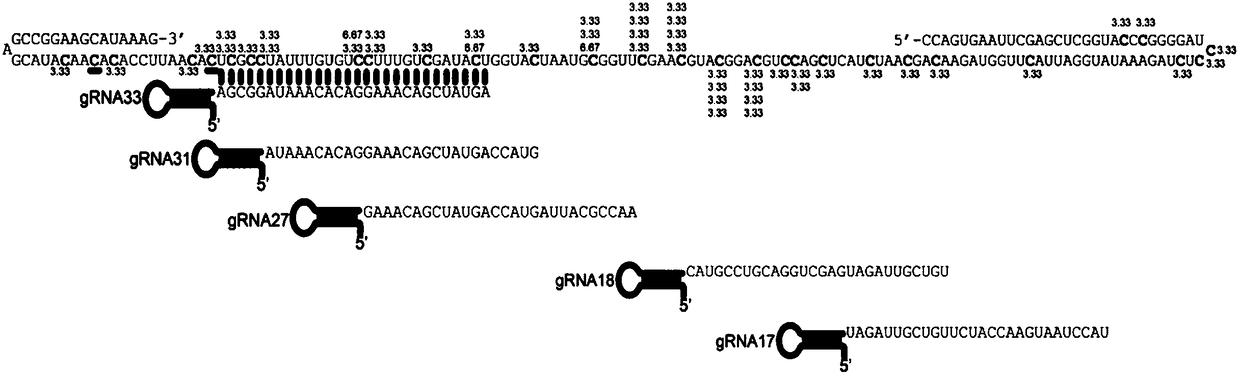
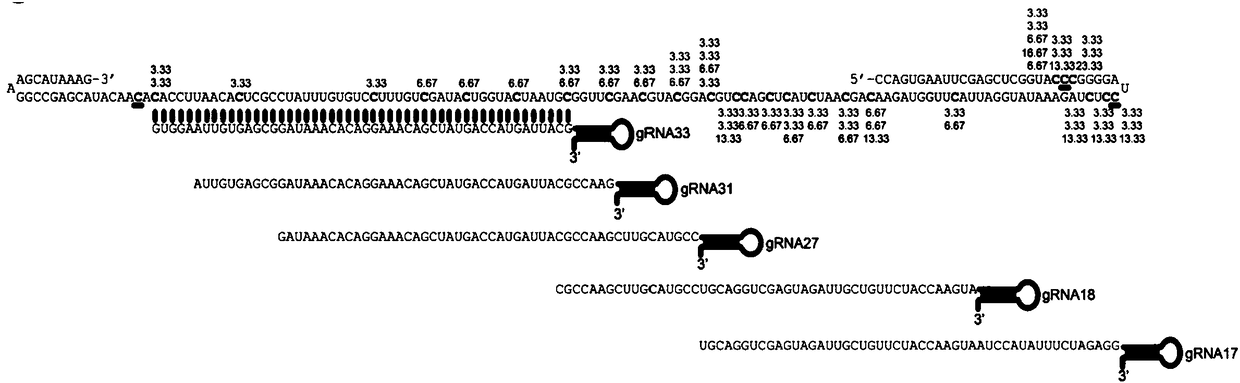
[0183] First, the expression vector was constructed and purified to obtain the fusion protein dCas13a-RsmB. The expression of dCas13a-RsmB was driven by the T7 promoter, and the Msb tag was added at the N-terminus to enhance expression, and there were His tags at the N-terminus and C-terminus for Purification of the fusion protein ( Figure 1A and Figure 1B ). In vitro methylation experiments were then performed, and the in vitro methylated RNA was treated with sodium bisulfite. The results of in vitro methylation of dCas13a-RsmB were obtained by sanger sequencing. Five gRNAs were used to target different positions of the target RNA. All five gRNAs used could methylate the target RNA, but different gRNAs The location and efficiency of methylation are different ( Figure 1C ). Although the in vitro methylation efficiency was low, ranging from 3.33% to 6.67%, dCas13a-RsmB could indeed mediate the methylation of cytosin...
Embodiment 2
[0184] Example 2 dCas13a-RsmB methylation in vivo
[0185] After successfully methylating RNA cytosine in vitro, try to modify RNA methylation in vivo. In order to achieve targeted RNA methylation in vivo, the subcellular localization of the Arabidopsis methyltransferase TRM4B was first performed. Figure 2AAs shown, TRM4B has only one conserved domain RsmB, so in the subsequent in vivo methylation experiments, RsmB is used instead of the full-length TRM4B for methylation experiments. The results of transient transformation of tobacco showed that TRM4B localized in the nucleus in planta ( Figure 2B and Figure 2C ), so in order to successfully carry out methylation in vivo, dCas13a-RsmB was introduced into the nucleus using nuclear localization signals. Then AtU6 was used to drive the expression of gRNA, and 35S was used to drive the expression of dCas13a-RsmB ( Figure 1A and Figure 4 ). The mRNA of MAG5 is the target RNA of TRM4B in Arabidopsis, and TRM4B can methylat...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap