A method for high-throughput screening of differentially expressed genes in plant genomes
A high-throughput, genomics technology, applied in the field of genetics research, can solve the problems of research limitations, Sanger sequencing can not further expand parallelism and microquantification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Raw material acquisition: Populus tomentosa '1316' is an excellent clone of Populus tomentosa from the National Germplasm Bank of Populus tomentosa;
[0064] All reagents used in the CTAB method are commercially available;
[0065] The specific operation steps are as follows:
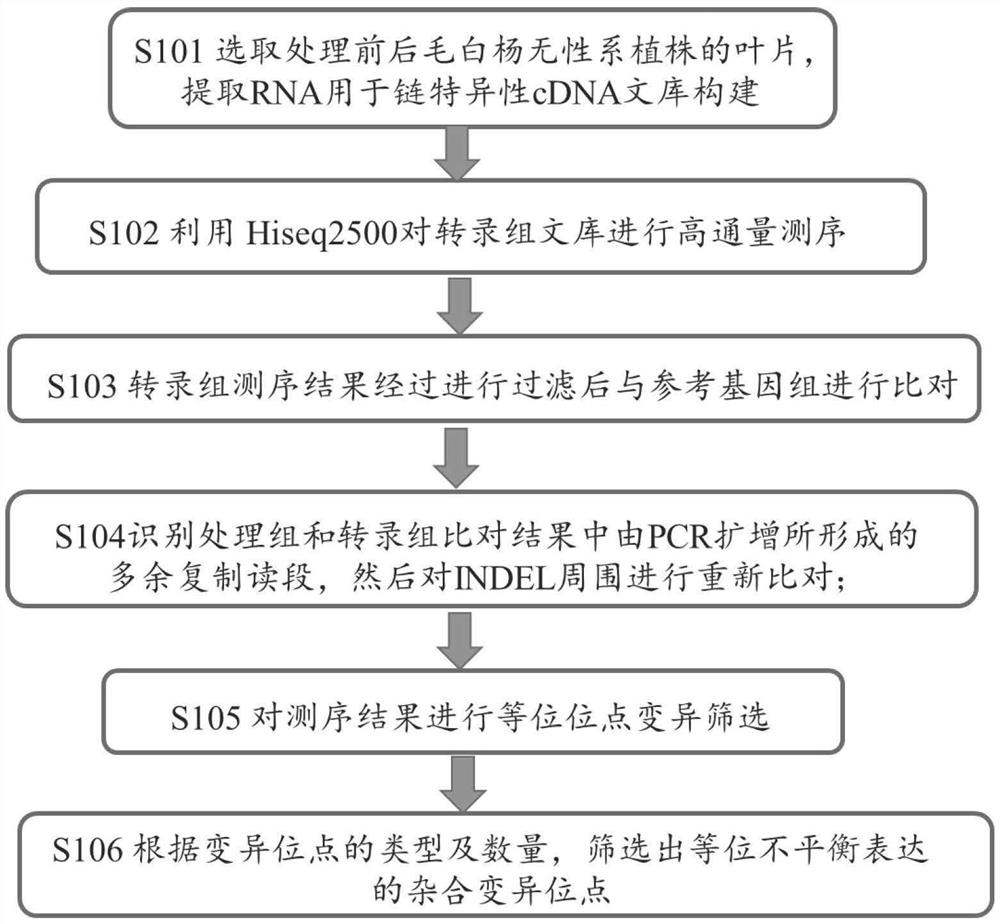
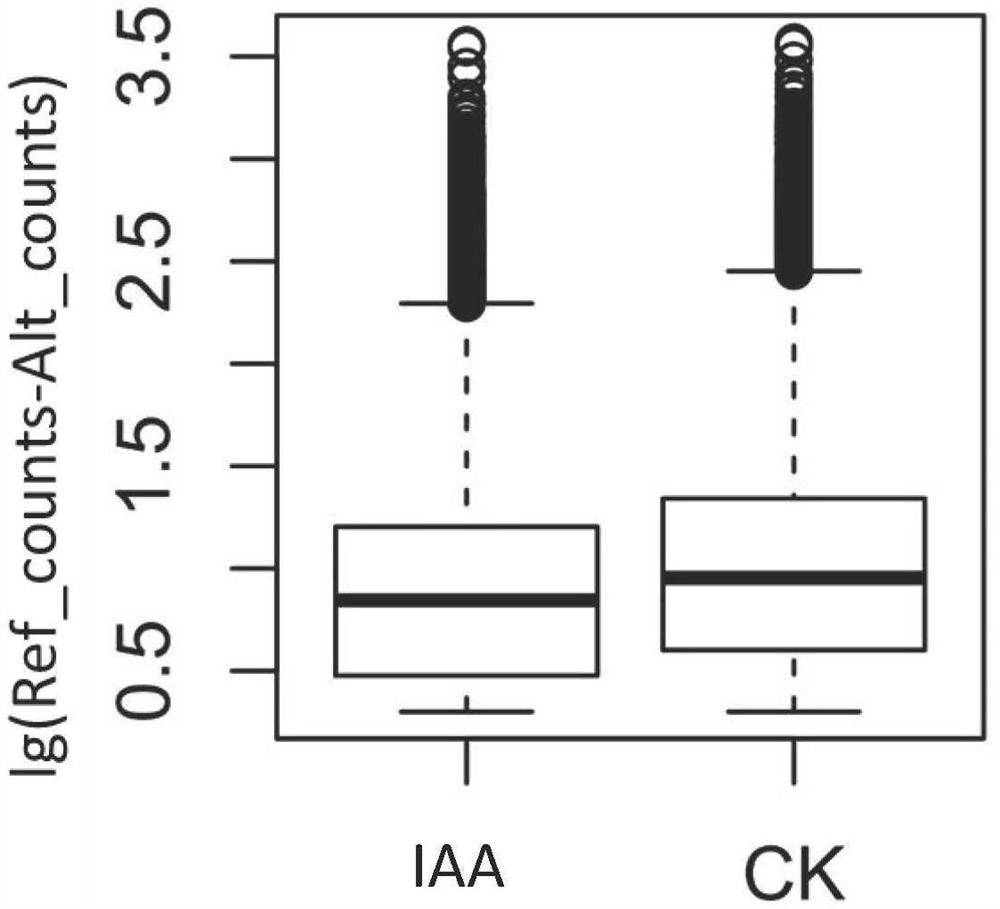
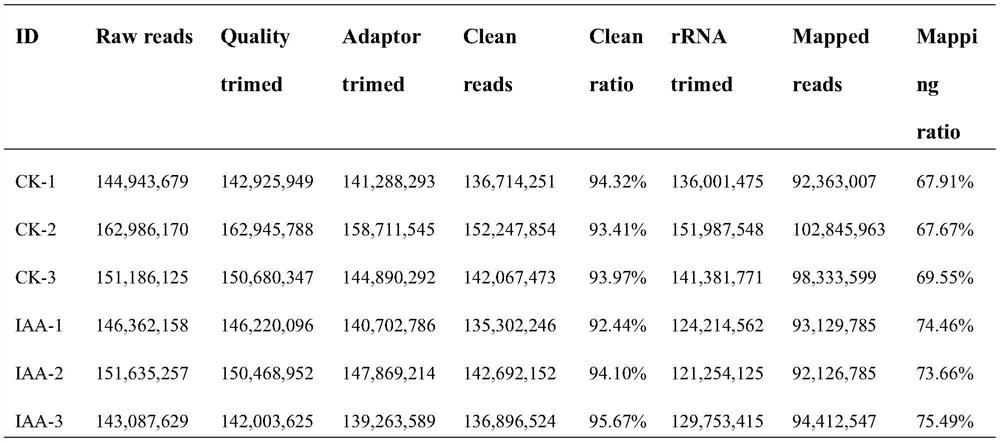
[0066]Step S101, select the annual plants of excellent clones of Populus tomentosa, and plant the clone plants used in the experiment in flower pots with soil, peat and perlite (volume ratio of 1:1:1) as the substrate, and the place is Beijing Forestry University Greenhouse (40°0'N, 116°20'E), the light time is 16 hours per day, and the temperature is 20°C. Adopt the IAA solution of 100 μ M (IAA powder is dissolved in 95% ethanol, then constant volume to 100 μ M with distilled water.) The leaves of the Populus tomentosa clone plant of the experimental sample are sprayed until the liquid drips on the leaves; the control sample is sprayed with the same Method of spraying with water. The leaves o...
Embodiment 2
[0108]The following will take the screening process of differential expression of alleles before and after drought stress treatment (taken from the National Populus microphylla germplasm resource bank) as an example to further illustrate the screening method provided by the present invention.
[0109] The acquisition of raw materials: select individuals with the same or similar growth potential in the Populus small-leaved clones, and carry out the following treatments: drought treatment, no watering for 10 days; control samples, watering according to the normal cultivation time (watering once every two days, until the soil is fully drenched).
[0110] All reagents used in the CTAB method are commercially available;
[0111] The specific operation steps are as follows:
[0112] Step S101, select excellent clone plants of Populus microphylla, and plant the clone plants used in the experiment in flower pots with soil, peat and perlite (1:1:1 volume ratio) as substrates, and the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com