A capture probe for detecting gene mutation by high-throughput sequencing and its application
A capture probe, high-throughput technology, applied in the field of capture probes for high-throughput sequencing detection of gene mutations, can solve the problems of reduced library richness, large probe usage, low capture efficiency, etc., to avoid capture efficiency and The effect of reducing probe utilization, ensuring capture efficiency, and high capture efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
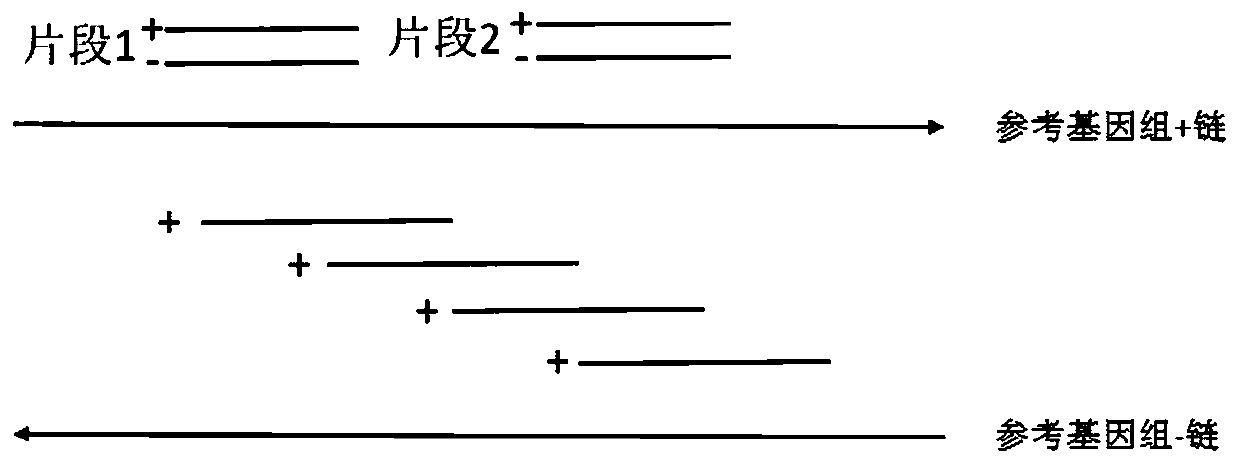
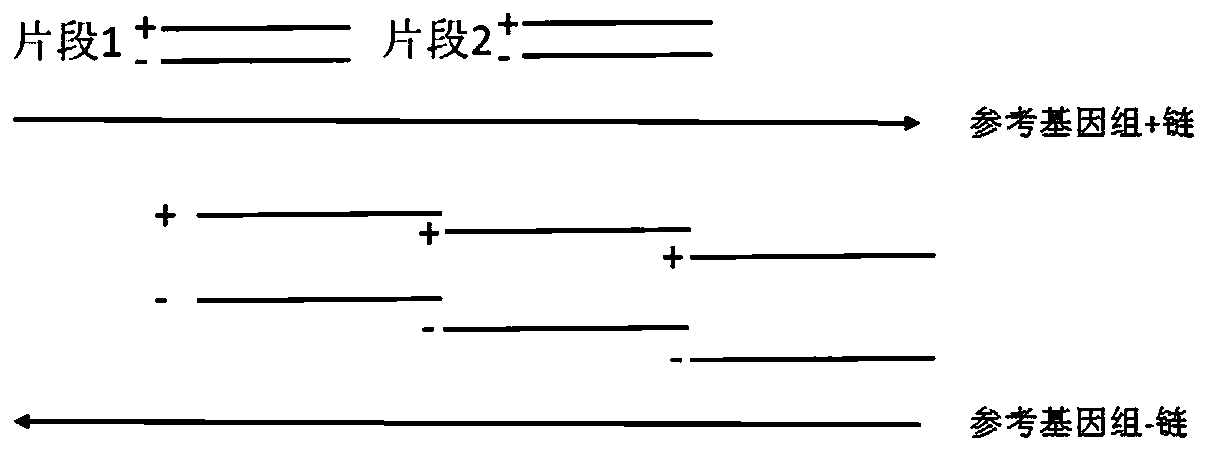
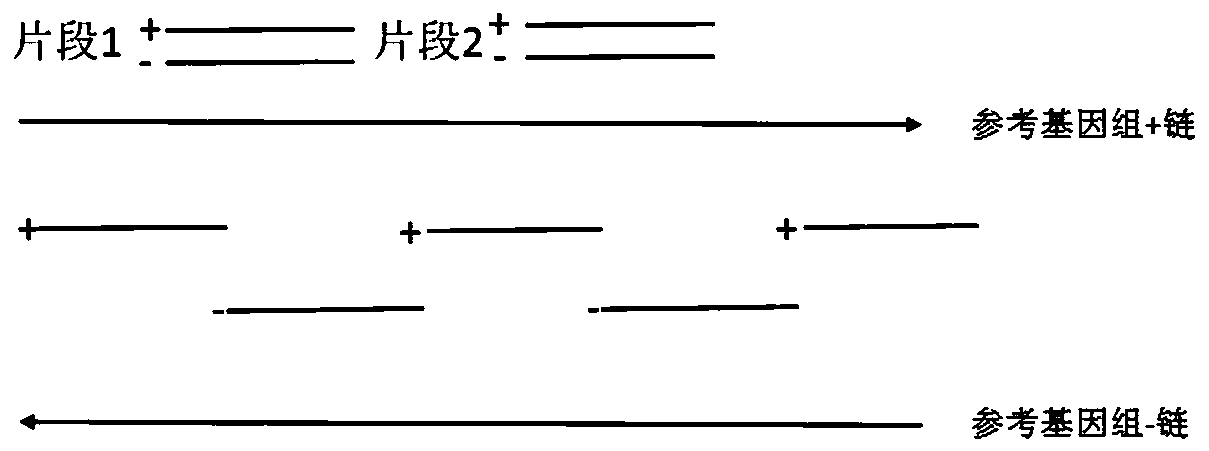
[0038] In this example, the epidermal growth factor receptor EGFR is taken as an example to verify the design method of the capture probe provided by the present invention. Three types of probes are designed for the distribution of the whole exon region of EGFR: traditional single-stranded probe (control group 1), double-stranded probe (control group 2) and sense strand probe and antisense strand provided by the present invention The probes are staggered single-stranded probes (experimental group), and the human genome library construction and the sequencing analysis of the EGFR hybridization capture library are carried out, and the library capacity, capture efficiency, repetition rate, and effective data depth of the constructed library are analyzed to compare different The capture effect of the capture probe.
[0039] 1. Probe design
[0040] (1) The traditional single-stranded probe design method is adopted to design single-stranded probes, and the single-stranded probes t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com