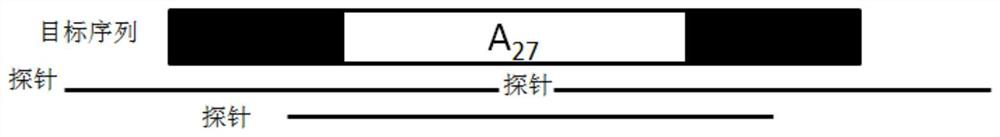
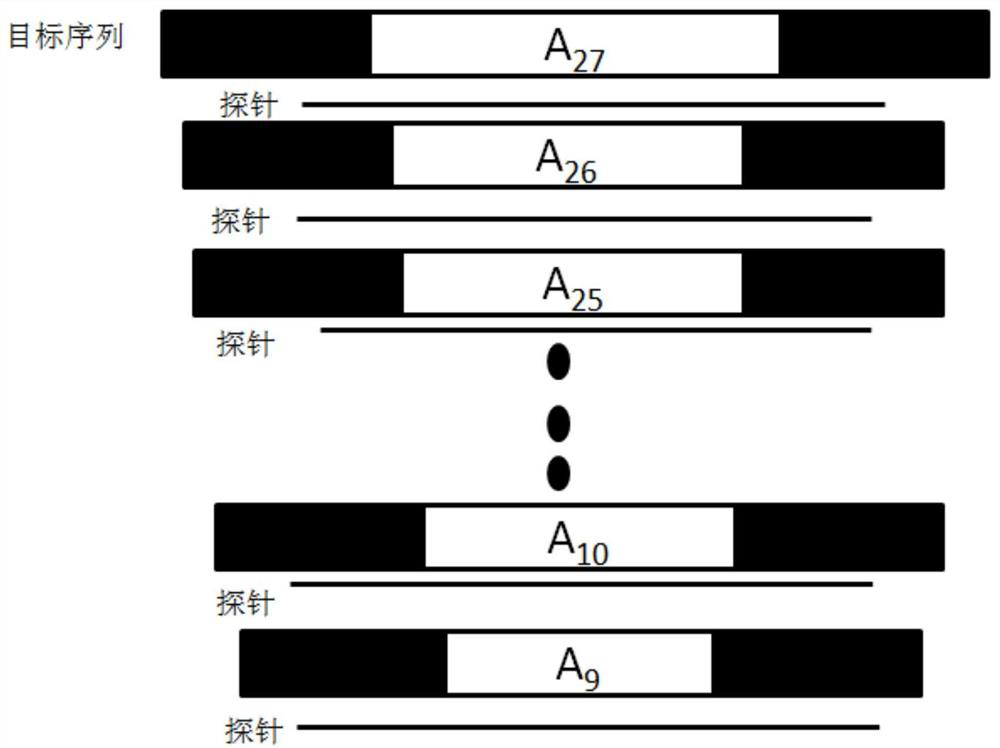
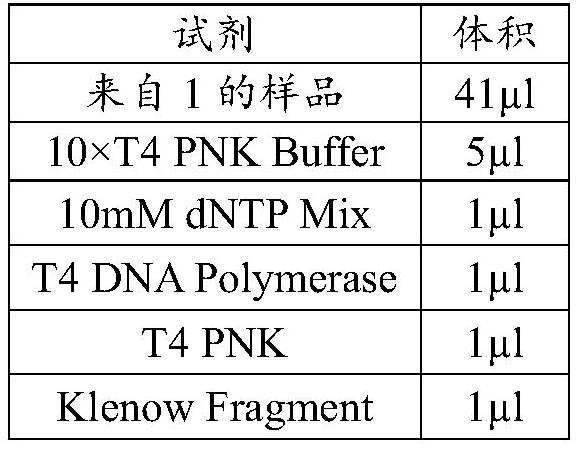
Probe set and reagent kit for detecting microsatellite instability through high-flux sequencing, and detection method of microsatellite instability
A microsatellite instability and detection method technology, applied in the field of detection of microsatellite instability, can solve the problems of genome instability and increased susceptibility to tumors of patients, and achieve the requirements of controlling sequencing costs, high effective depth, and reducing sample size Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] The preparation of embodiment 1 capture probe
[0050] Probe sequences were synthesized using methods known in the art, uniformly mixed in TE with a total volume of 1 ml, and 1 μl of them were used for PCR amplification using the general PCR primers shown in the following table. PCR amplification system: the above-mentioned probe solution, 1 μl; Forward primer (20 μM), 1 μl; reverse primer (20 μM), 1ul; 47 μL Platinum PCR Supermix, total volume 50 μL. Amplification conditions: 95°C, 2min; (94°C, 30s, 55°C, 30s, 72°C, 30s) 20 cycles; 72°C, 5min.
[0051] Sequence (5'-3') TAATACGACTCACTATAGGGATCGCACCAGCGTGT AGAAGAGTCACATAC
[0052] The PCR product was purified with MinElute PCR Purification Kit and stored at -20°C. Take 500ng, and use Ambion SP6 megascript kit to perform in vitro transcription on the product after PCR purification. The in vitro transcribed RNA was purified using Qiagen RNeasy minikit, and the purified product was the biotin-labe...
Embodiment 2
[0053] Example 2 Specific capture and sequencing
[0054] In this example, the capture probe set provided in Example 1 was used, and the target sequence was enriched by the sequence capture technology based on liquid phase hybridization. The technical route of liquid phase hybridization is: 1) preparing a hybridization probe library, 2) using the probes to enrich the target gene, and 3) sequencing the enriched DNA sequence with a high-throughput sequencer.
[0055] The library construction method comprises the following steps:
[0056] 1. DNA extraction and fragmentation
[0057] DNA was extracted from 4 peripheral blood samples using the QIAamp DNA Blood Mini Kit.
[0058] Use the Bioruptor Pico DNA interrupter, after the temperature of the cold cycler drops to 4°C, set the parameters ON for 30s, OFF for 30s as a cycle, every 10 cycles as a round, a total of 3 rounds, after each group, the samples were placed in Mix well on a shaker, centrifuge briefly and interrupt for th...
Embodiment 3
[0100] 20 colorectal cancer samples with known MSI types and plasma samples from corresponding patients were collected, and tested according to the procedure shown in Example 2. The test results are shown below.
[0101]
[0102] Can draw from above-mentioned result, use probe of the present invention to detect microsatellite state of different samples, the accuracy of tissue sample reaches 100%, specificity reaches 100%, the accuracy of plasma sample reaches 97.5%, specificity reaches 100% %.
[0103] According to the above description, the present invention has the following advantages: 1. High accuracy, strong sensitivity, high throughput, and can effectively and accurately detect microsatellite instability in tumor patients. 2. The microsatellite instability detection method based on next-generation sequencing data can eliminate the need for a separate and specialized detection of MSI status, and can detect microsatellite instability in clinical practice while detecting...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com