A recombinant fusion protein that specifically recognizes tumor cells and its application
A fusion protein and tumor cell technology, applied in the field of recombinant fusion protein, can solve the problems of high cost, high operation threshold of colorectal cancer early diagnosis technology, lack of genetic engineering means for specific identification of intestinal tumors, etc. Easy to use effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Construction of plasmid bacterial surface display pCDFDuet1-INP-tPep of
[0040]Plasmid pET-InaZN-EGFP (S.Bao, S.Yu, X.Guo, et al.Journal of AppliedMicrobiology, 2015 (119):. 236-244) as a template and primers designed and SacI-INPF HindIII-tPep- INPR PCR amplification, to obtain a DNA fragment of INP-tPep. Wherein an upstream primer SacI-INPF as: CGAGCTCCTGTACCATGGATCTCGACAA (SEQ ID NO: 1); downstream primer HindIII-tPep-INPR as: CCCAAGCTTGGGTTAAATACGAGAAGCAGGAGACCAAGCATACCAACGATGCCAAACAATCGTGGATCCGTAAGTGG (SEQID NO: 2). Further by the SacI and HindIII double enzyme digestion, DNA fragments having cohesive ends. Which was constructed between the enzyme-linked by the process of digestion of plasmid pCDFDuet1 (available from Novagen, 71340-3 Num) of SacI and Hindlll cloning site, forming plasmid pCDFDuet1-INP-tPep with INP-tPep of transform E. coli, using streptomycin screened to obtain transformed strains. After plasmid extraction and purification, DNA sequencing, the plasm...
Embodiment 2
[0042] Construction of recombinant bacteria INP-tPep-EGFP
[0043] Construction of pGEX-4T-1-EGFP plasmid: by PCR from the plasmid pEGFP-N1 (preferably available from Takara Bio) as a template, amplified EGFP-FP with the following primers: GCGGATCCATGGTGAGCAAGGGCGAGGAG (SEQ ID NO: 6); EGFP- RP: GCGAATTCTTACTTGTACAGCTCGTCCATGCCG (SEQ ID NO: 7) product recovery after double digestion with BamHI and EcoRI, connected to the same double digestion of pGEX-4T-1 plasmid (preferably available from Takara bio), transformation, positive clones were obtained by screening ampicillin . After by cotransformation The plasmid pCDFDuet1-INP-tPep plasmid pGEX-4T-1-EGFP was transformed into E. coli BL21 (genotype F-ompThsdS (rB-mB-) galdcm (DE3)), the expression allowed. Specific steps are as follows:
[0044] 1) Transformation: each 5μl pCDFDuet1-INP-tPep with pGEX-4T-1-EGFP plasmid was added 50μl of E. coli competent cells, placed on ice for 30 minutes, 42 ℃ heat shock 90s, then returned to ice for...
Embodiment 3
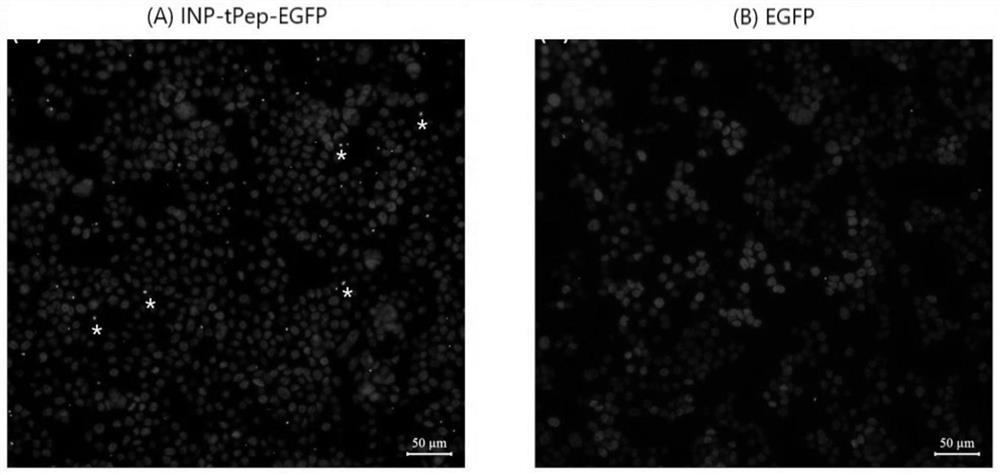
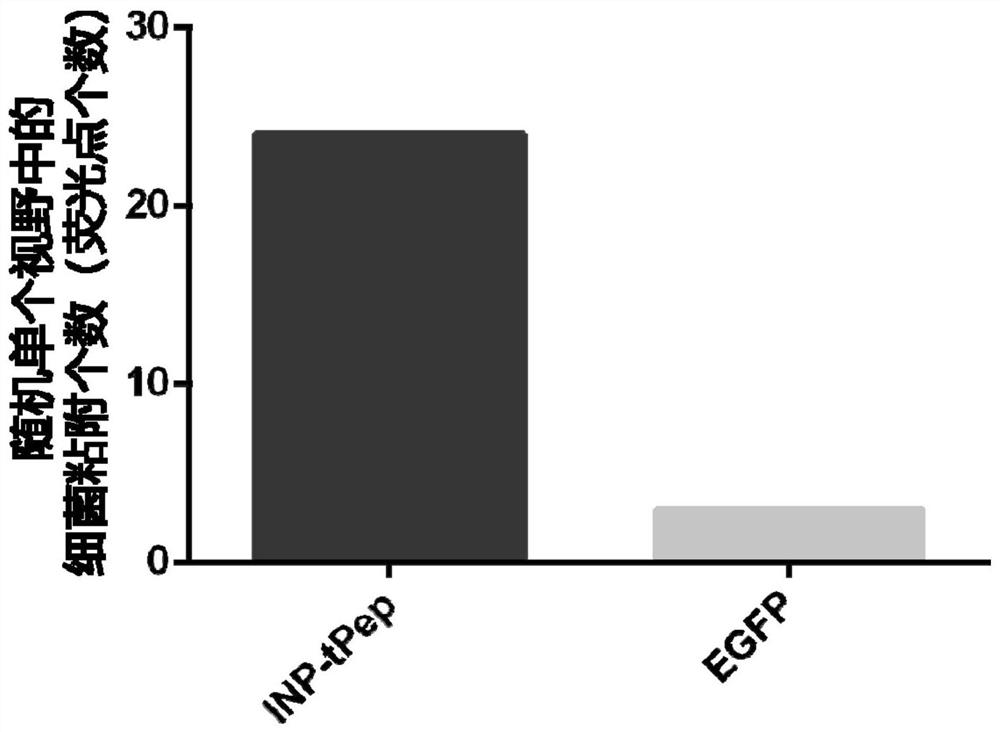
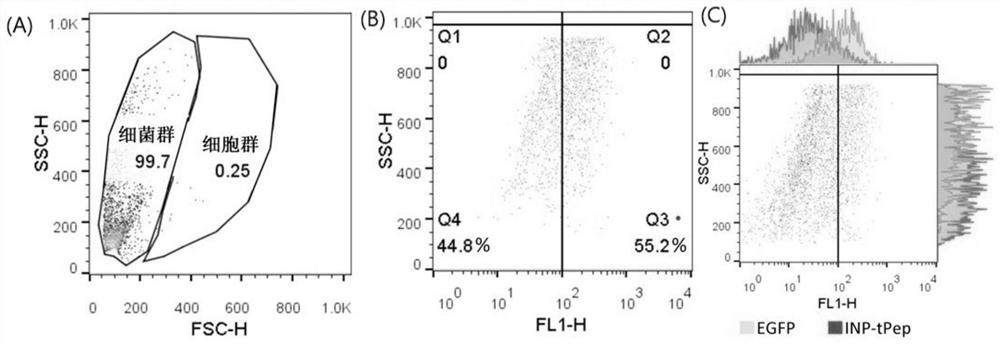
[0048] Cells climbing film identified recombinant bacteria INP-tPep-EGFP and human HT29 colorectal cancer cell specifically recognizing placed 2cm diameter climbing film cells in 24-well cell culture plate, each well 0.5ml of 5% of the density of HT29 cells; cell proliferation when the rear cover 70% to 80% of the slide, the medium was changed to normal saline (0.5ml / well) was added 100μlINP-tPep-EGFP EGFP recombinant bacterium or bacteria (i.e., only into the bacterial pGEX-4T-1-EGFP plasmid) was (OD600≈1.0), placed in a 37 [deg.] C shaker were incubated 0.5 hours after incubation, cells were removed climbing film, with a 4% paraformaldehyde fixed cells, and treated with nuclei were stained DAPI staining solution; DAPI fluorescence microscope at 488 and climbing film passage cells were photographed, and the combined image of two channels. Such as figure 1 Indicated (shown) figure 1 A is INP-tPep-EGFP recombinant bacterial binding to cells; figure 1 B is EGFP recombinant bacteri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com