A simple and efficient method for protein purification
A protein and protein purification technology, applied in the field of bioengineering, can solve problems such as high cost, target protein damage, and limited application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Purification of enhanced green fluorescent protein eGFP
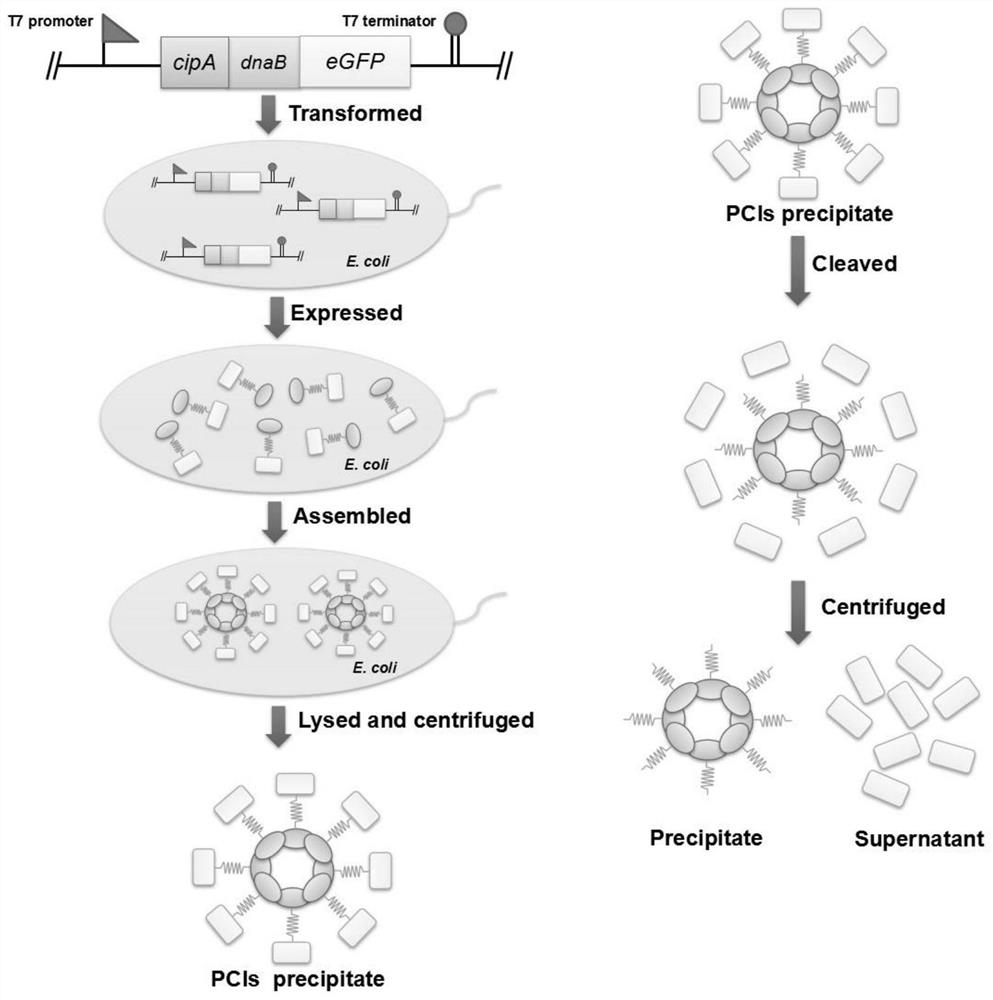
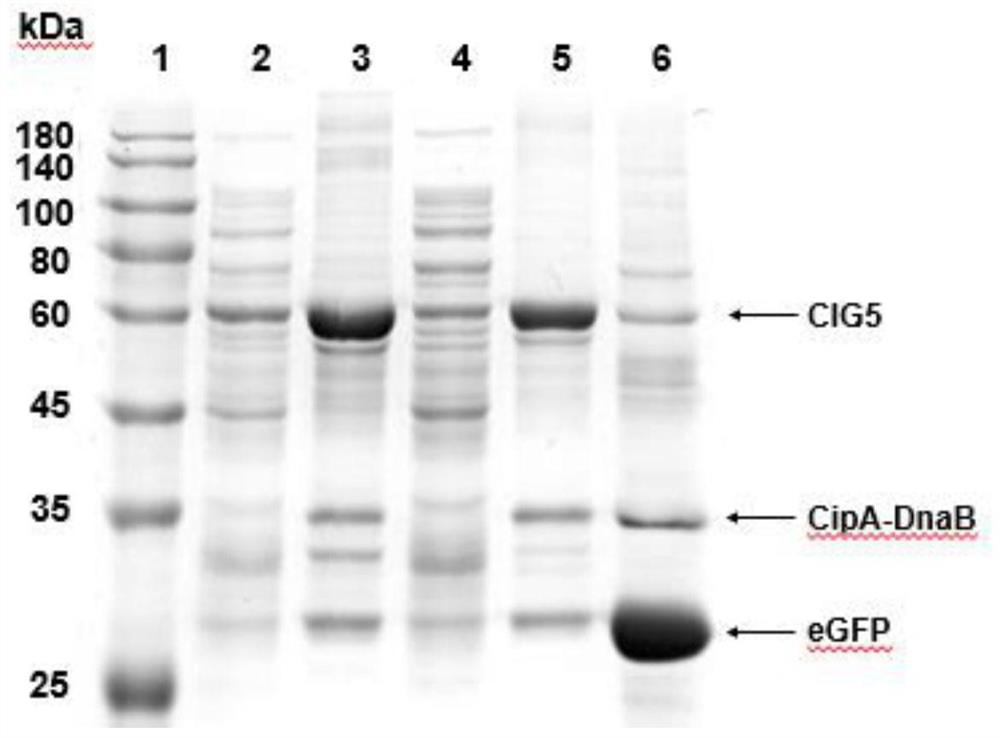
[0051] (1) The plasmid pET-28a-cipA-dnaB-eGFP was constructed, and the genes cipA (NCBI: CAE138691), dnaB (PDB: 1MI8_A) and eGFP (NCBI: AHK23750.1) were synthesized by OE-PCR. Ligate cipA and eGFP to the 5' and 3' ends of dnaB, respectively. cipA-dnaB-eGFP was cloned into pET-28a vector by Gibson assembly. Subsequently, the PCR product was transformed into E. coli XL10-Gold, and the plasmid was verified by DNA sequencing.
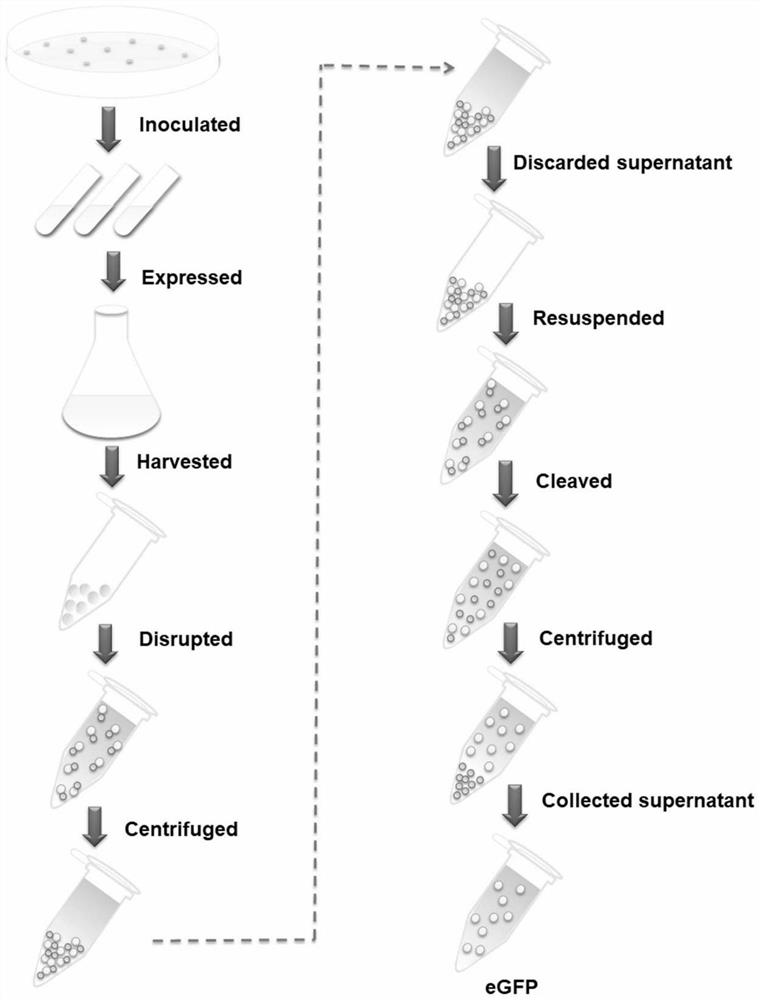
[0052] (2) Express the protein CipA-DnaB-eGFP, introduce the plasmid pET-28a-cipA-dnaB-eGFP into Escherichia coli BL21(DE3), pick a single colony and inoculate it in 5mL LB medium, add 50μg / mL Kana Seed solution was obtained by culturing overnight at 37°C. The next day, the seed solution was inoculated in 200mL fresh TB medium, and cultured at 37°C for about 2h, so that the OD 600 When it reaches 0.5-0.8, add 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) to the culture medium, and culture at...
Embodiment 2
[0058] Purification of β-galactosidase (β-Gal)
[0059] (1) The plasmid pET-28α-cipA-dnaB-lacZ (pET-28α-CIZ) was constructed, and the genes cipA (NCBI: CAE138691), dnaB (PDB: 1MI8_A) and lacZ (NCBI: 945006) were synthesized by OE-PCR . Then, cipA and lacZ were ligated to the 5' and 3' ends of dnaB by OE-PCR, respectively. The cipA-dnaB-lacZ fragment was then cloned into the pET-28a vector by Gibson assembly. Subsequently, the PCR product was transformed into Escherichia coli XL10-Gold, and the recombinant plasmid was verified by sequencing.
[0060] (2) Express the fusion protein CipA-DnaB-β-Gal, transform pET-28α-CIZ into Escherichia coli BL21(DE3), and inoculate a single colony in 5 mL LB medium, add 50 μg / mL kanamycin , inoculate the culture solution in 200 mL of fresh TB medium after culturing overnight at 37 °С. OD 600 When it reaches 0.5-0.8, add 0.5mM isopropyl-β-D-thiogalactoside (IPTG) and incubate at 30°C for 10h. In order to prevent the acidification of the me...
Embodiment 3
[0069] Purification of Maltose Binding Protein (MBP)
[0070] (1) The plasmid pET-28α-cipA-dnaB-MBP (pET-28α-CIM) was constructed, and the genes cipA (NCBI: CAE138691), dnaB (PDB: 1MI8_A) and MBP (Gene ID: 4155 ). Then, cipA and MBP were ligated to the 5' and 3' ends of dnaB by OE-PCR, respectively. The cipA-dnaB-MBP fragment was then cloned into the pET-28a vector by Gibson assembly. Subsequently, the PCR product was transformed into Escherichia coli XL10-Gold, and the recombinant plasmid was verified by sequencing.
[0071] (2) Express the fusion protein CipA-DnaB-MBP, transform pET-28α-CIM into Escherichia coli BL21 (DE3), and inoculate a single colony in 5 mL LB medium, add 50 μg / mL kanamycin, in After culturing at 37°C overnight, the culture solution was inoculated into 200 mL of fresh TB medium. OD 600 When it reaches 0.5-0.8, add 0.5mM isopropyl-β-D-thiogalactoside (IPTG) and incubate at 30°C for 10h. In order to prevent the acidification of the medium in the medi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com